CPDadh: A new peptidase family homologous to the cysteine protease domain in bacterial MARTX toxins
Abstract
A cysteine protease domain (CPD) has been recently discovered in a group of multifunctional, autoprocessing RTX toxins (MARTX) and Clostridium difficile toxins A and B. These CPDs (referred to as CPDmartx) autocleave the toxins to release domains with toxic effects inside host cells. We report identification and computational analysis of CPDadh, a new cysteine peptidase family homologous to CPDmartx. CPDadh and CPDmartx share a Rossmann-like structural core and conserved catalytic residues. In bacteria, domains of the CPDadh family are present at the N-termini of a diverse group of putative cell-cell interaction proteins and at the C-termini of some RHS (recombination hot spot) proteins. In eukaryotes, catalytically inactive members of the CPDadh family are found in cell surface protein NELF (nasal embryonic LHRH factor) and some putative signaling proteins.
Introduction
RTX (repeats-in-toxin) toxins refer to a diverse group of large proteins secreted by Gram-negative bacteria, including Escherichia coli α-hemolysin, Pasteurella haefflolytica leukotoxin, and Bordetella pertussis adenylate cyclase toxin.1 They are characterized by repeats of a glycine and aspartate-rich, calcium-binding sequence motif. Recently, a family of multifunctional, autoprocessing RTX toxins (MARTX), typified by VcRtxA from Vibrio cholerae, are found to contain a cysteine protease domain (CPD, referred to as CPDmartx in this work). CPDmartx autocleaves these toxins to release domains with toxic effects to the cytosol of host cells.2 This domain is also used in the autocleavage of toxins A and B from the Gram-negative, pathogenic Clostridium difficile.3 Here, we report identification and computational analysis of a new cysteine peptidase family homologous to CPDmartx in a diverse group of bacterial proteins and their homologs in eukaryotes with potentially lost peptidase activity.
Results and Discussions
Identification of a new family of cysteine protease domains (CPDadh) homologous to CPDmartx
PSI-BLAST4 searches (see Materials and methods) for CPDmartx domains converge to about 70 bacterial proteins. Identified sequences display relatively high similarity to each other. To investigate if remote homologs of CPDmartx domains exist, we manually inspected PSI-BLAST hits above the default e-value cutoff (0.001). The N-terminal region of a large protein annotated as “Autotransporter adhesion” from Magnetospirillum gryphiswaldense [NCBI gene identification (gi) number 144897667, with an e-value of 0.68] was identified to have two sequence motifs of CPDmartx that harbor the conserved histidine and cysteine catalytic diad.5 A PSI-BLAST search starting from this domain (gi|144897667, residues 88–256) revealed that most of its bacterial homologs also have the conserved histidine and cysteine residues in these motifs. Comprehensive PSI-BLAST searches starting from multiple representatives of found homologs of gi|144897667 identified about 300 proteins containing this new domain.
Regions corresponding to this new domain do not contain known domains as revealed by submitting found proteins to domain databases such as CDD,6 Pfam7 and SMART.8 CPDmartx domains and the new family of domains could not find each other as significant hits during PSI-BLAST searches, suggesting limited sequence similarity between them. Similarity searches against the pdb70 database (protein databank9 nonredundant sequences with known structures at 70% identity) using a more sensitive profile–profile comparison method HHpred10 did suggest this new domain is remotely related to the CPDmartx domain of V. cholerae with a recently solved structure (pdb id: 3eeb; HHpred probability of 0.77).5 We also submitted this new domain to the 3D-Jury Meta server,11 which assembles the results of various fold recognition methods and computes consensus scores for the predictions. Several fold recognition methods, such as Meta-Basic,12 FFAS0313 and mGenThreader,14 found CPDmartx structure 3eeb and/or other structures with the caspase-like fold as top hits. The best hit of the 3D-Jury consensus results is 3eeb with a significance score above 60. Other structures with the caspase-like fold were also among the top consensus hits; and the conserved cysteine and histidine residues in this new domain are aligned to the corresponding catalytic residues in these structures. These results indicate that this new domain has a caspase-like fold and is homologous to CPDmartx.
Few proteins with this new domain have been experimentally characterized. Annotations for many of them are simply hypothetical proteins or based on other domains present in them, such as “hemolysin-type calcium-binding region” (gi|158520629), “hemagglutination activity domain-containing protein” (gi|158341333) and “putative outer membrane adhesin-like protein” (gi|119946789). As co-occurring domains are often involved in cell adhesion, we refer to this new family of putative cysteine protease domains as CPDadh.
Sequence and structure characterization of CPDadh domains
In the MEROPS peptidase classification database,15 CPDmartx domain is denoted as peptidase family C80 in clan CD. This clan also includes several remotely related peptidase families, such as clostripain (C11), legumain (C13), caspase (C14), gingipain (C15), and separase (C50). Comprehensive sequence similarity searches and evolutionary analyses were conducted several years ago for peptidases in this clan.16 The structures of caspase17 and gingipain18 have been well characterized, both having a Rossmann-fold like core with a mainly parallel beta sheet surrounded by alpha-helices on both sides. Compared to caspases, the most noticeable differences in the structure of recently solved CPDmartx from V. cholerae5 lies in the C-terminus, where two helices of caspases are replaced by several beta strands in CPDmartx. These beta strands form part of the binding pocket for the small molecule inositol hexakisphosphate (InsP6).5
Multiple sequence alignment19 and secondary structure predictions20 reveal that most CPDadh domains retain the core structure elements characteristic of caspases or CPDmartx (see Fig. 1). The exception are for bacterial group 2 CPDadh domains (sequence grouping is discussed below), where the N-terminal regions before the beta-strand preceding the active site histidine are quite diverse (not shown in Fig. 1). The C-terminal ends for these proteins are also divergent (not shown in Fig. 1). As the C-terminus of the V. cholerae MARTX CPD is involved in protease activation in response to InsP6, it is possible that the divergent C-termini of CPDadh domains have evolved responsiveness to other small molecule or protein ligands to allow for activation in diverse environments. Indeed, the basic residues involved in InsP6 binding in the V. cholerae MARTX CPD are not conserved in CPDadh domains, indicating different mechanism(s) of activation.
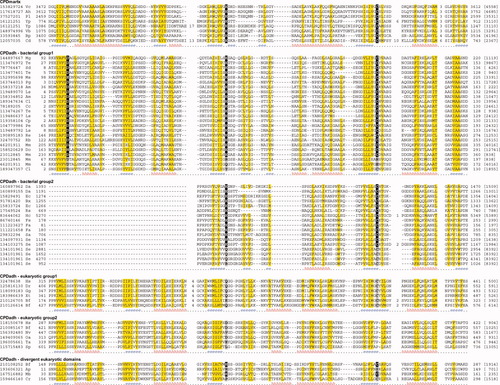
Multiple sequence alignments of CPDmartx and CPDadh domains. Nonpolar residues in positions with mainly hydrophobic residues are shaded in yellow. Catalytic residues are shaded in black with the exception of putative inactive members where they are shaded in gray. Starting and ending residues numbers, as well as sequence lengths (in brackets), are shown. Long insertion regions in the alignment are replaced by the numbers of residues. Consensus secondary structure predictions are shown below alignment of each group (h, helix; e, strand). The proteins are identified by their NCBI gene identification (gi) numbers, followed by the species name abbreviations: Ah, Aeromonas hydrophila; Am, Acaryochloris marina; Ap, Acyrthosiphon pisum; Bf, Branchiostoma floridae; Bp, Bordetella pertussis; Bx, Burkholderia xenovorans; Cc, Chlorobium chlorochromatii; Cd, Clostridium difficile; Cl, Chlorobium limicola; Cp, Chlorobium phaeobacteroides; Cr, Chlamydomonas reinhardtii; Da, Delftia acidovorans; Do, Desulfococcus oleovorans; Dr, Danio rerio; Ec, Escherichia coli; Fa, Frankia alni; Fs, Frankia sp.; Gg, Gallus gallus; Hc, Hahella chejuensis; Hs, Homo sapiens; Ls, Lyngbya sp.; Mb, Monosiga brevicollis; Mg, Magnetospirillum gryphiswaldense; Mm, Magnetospirillum magnetotacticum; Ms, Marinomonas sp.; Nv, Nematostella vectensis; Oa, Ornithorhynchus anatinus; Pe, Pseudomonas entomophila; Pg, Phaeobacter gallaeciensis; Pl, Photorhabdus luminescens; Rb, Rhodopirellula baltica; Re, Rhizobium etli; Rs, Ralstonia solanacearum; Sa, Streptomyces avermitilis; Sd, Sulfurimonas denitrificans; Se, Saccharopolyspora erythraea; Sp, Strongylocentrotus purpuratus; Ta, Trichoplax adhaerens; Te, Trichodesmium erythraeum; Vb, Vibrionales bacterium; Vc, Vibrio cholerae; Xt, Xenopus tropicalis; Yp, Yersinia pestis.
Grouping and domain contents of CPDadh domain-containing proteins
CPDadh domains are found in proteins from bacteria and eukaryotes. Most of the bacterial CPDadh-containing proteins are long, with more than one thousand residues. They have a much wider species distribution than CPDmartx-containing proteins, which are only present in several pathogenic proteobacteria and Clostridium species in current sequence database. The CLANS program21 was used to cluster CPDadh domains based on BLAST4 scores and display the results (Supporting Information Fig. 1). The results suggest there are mainly two bacterial groups and two eukaryotic groups with distinct sequence annotations and domain contents (see Fig. 2).
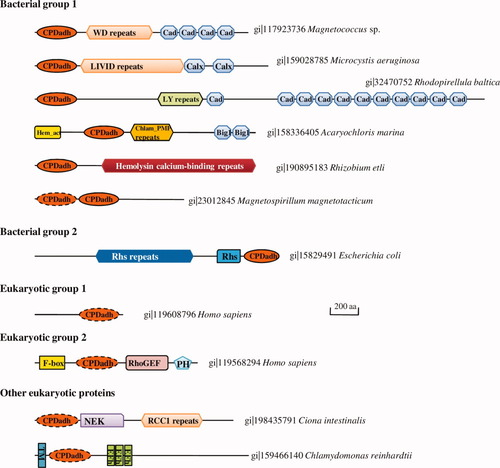
Domain architecture of selected CPDadh-containing proteins. Regions containing various repeats are marked with hexagons. Domains with immunoglobulin fold are shown as octagons. Domain name abbreviations are: Cad, cadherin domain; Calx, Calx-beta domain; Hem-act, haemagglutination activity domain; TM, transmembrane region; EF, EF-hand calcium binding domain. Putative catalytically inactive CPDadh domains have dashed outlines.
Bacterial CPDadh-containing proteins
Bacterial group 1
This is the largest group with about 140 proteins. Annotations of these proteins are usually based on domain contents or simply hypothetical proteins. Although few experimental studies have been carried out on these proteins, most of them probably function as cell adhesion molecules as the co-occurring domains are mostly involved in cell surface protein-protein interactions. The domain contents of these proteins are diverse (Fig. 2), as revealed by analysis using the hmmpfam program against the latest Pfam database7 (see supplementary hmmpfam results at http://prodata.swmed.edu/CPDadh). Many domains are themselves repeats or have multiple copies in one protein. The repeats often adopt beta-roll structures such as hemolysin-type calcium-binding repeat (Pfam family PF00353)22 and pentapeptide repeat (PF00805)23; or beta-propeller structures such as WD40 repeat,24 BNR/Asp-box repeat (PF02012),25 FG-GAP repeat (PF01839)26 and LIVID repeat (PF08309).27 Domains with immunoglobulin fold are abundant, such as cadherin domain (PF00028),28 PKD domain (PF00801),29 Calx-beta domain (PF03160),30 and peptidase family C25 C terminal ig-like domain (PF03785).31
The CPDadh domain is at the N-termini of these proteins, and is often the first domain. Some proteins (e.g., gi|89075996 and gi|90578280) have a signal peptide present at the N-terminus, suggesting that they are secreted via the Sec dependent secretion pathway. In a few cases, two tandem CPDadh domains occur at the N-terminus (e.g., gi|23012845, Fig. 2), suggesting domain duplication events. The common domain localization of CPDadh in these proteins suggests that it perform a function with similar mechanism. One possibility is that this domain could be involved in autoprocessing of these bacterial proteins. Besides the conserved catalytic histidine and cysteine residues, a conserved Asp/Asn residue is present at the end of the first core beta strand in CPDadh domains of bacterial group 1 proteins (see Fig. 1). This Asp/Asn residue could contribute to catalytic reaction or substrate binding. A few proteins have one or more active site residues mutated, possibly resulting in inactivation.
Using the STRING server32 to mine genomic contextual information, we found that some bacterial group 1 CPDadh-containing proteins have genomic neighbors that are components of type I secretion systems (T1SS),33 suggesting that they are secreted by T1SS. For example, the protein VCSB (Swiss-Prot id Q3B5W4) from Pelodictyon luteolum is associated with a transport ATPase and type I secretion membrane fusion protein HlyD. RTX toxins, including the CPDmartx-containing MARTX proteins, are also secreted by T1SS.2 Products of other genomic neighbors of bacterial group 1 CPDadh-containing proteins are sometimes annotated as outer membrane proteins and often contain domains involved in cell-cell interactions too, suggesting that these proteins function together to mediate cell adhesion.
The majority of bacterial species with this group of CPDadh domains are from the phyla of proteobacteria and cyanobacteria. They are mostly free-living, nonpathogenic bacteria from aquatic environment. A few cyanobacteria species, such as Lyngbya sp. and Acaryochloris marina, have more than 10 proteins with CPDadh domains.
Bacterial group 2
This group of CPDadh domains exhibits high sequence divergence (Supporting Information Fig. 1). Bacterial species with this group of CPDadh domains are mainly from the phyla of proteobacteria and actinobacteria. Many members are associated with the RHS (recombination hot spot) proteins.34 RHS proteins have been identified in various strains of E. coli, all of which have the characteristic RHS repeats (PF05593, also called YD repeats due to the conserved “YD” residues in repeated motif “xxGxxxRYxYDxxGRL[I/T]xxxx”). Other experimentally characterized RHS proteins include a cell wall associated protein in Bacillus subtilis,35 insecticidal toxins TccC from Photorhabdus luminescens36 and SepC from Serratia entomophila,37 and a class of eukaryotic transmembrane proteins called teneurins.38 The C-termini of different RHS proteins are divergent and can contain various nonhomologous domains. The CPDadh-containing RHS proteins, including two (RhsA and RhsG) from E. coli strain O157, all have the CPDadh domain located at the C-terminal end. One CPDadh-containing RHS protein from the nematode symbiont species Xenorhabdus bovienii is annotated to be toxic to nematodes (unpublished results, gi|11967898). The CPDadh domains in RHS proteins could contribute to the processing of these proteins, or act as a virulence activity domain. This group also includes proteins without RHS repeats, some of which (e.g., gi|134101961, Fig. 1) contain multiple copies of inactivated CPDadh domains.
Eukaryotic CPDadh-containing proteins
Two main groups of eukaryotic CPDadh domains exist with some divergent members. The active site histidine residues are mutated in most of eukaryotic proteins, suggesting loss of peptidase activity.
Eukaryotic group 1
This group contains proteins named NELF (nasal embryonic LHRH factor) in vertebrates. Their function is related to migration of LHRH (luteinizing hormone-releasing hormone) neurons during embryonic development.39 NELF is located on the outside of LHRH cell membrane and could be a cell adhesion molecule.39 Mutation of human NELF protein has been linked to Kallmann syndrome (hypogonadotropic hypogonadism with anosmia/hyposmia).40 NELF orthologs were found from sea urchin Strongylocentrotus purpuratus and placozoan Trichoplax adhaerens,41 suggesting its ancient origin in animals. CPDadh domains are located at the C-terminal ends of NELF proteins. N-terminal regions of these proteins do not have many regular secondary structures. As the active site histidine residues are mutated, CPDadh domains in NELF proteins probably do not have peptidase activity, but could contribute to cell adhesion though protein or peptide binding. The active site cysteine residues in these domains are preserved (see Fig. 1), and probably still play a role in the function of NELF proteins.
Eukaryotic group 2
This group consists of proteins annotated as hypothetical proteins or F-box containing proteins. The functions of these proteins are unknown. They contain an F-box domain,42 a RhoGEF (guanine nucleotide exchange factor for Rho/Rac/Cdc42-like GTPases) domain43 and a PH (pleckstrin homology) domain44 (see Fig. 2), suggesting possible roles in signaling pathways. These proteins are present in vertebrates as well as some lower animals such as Trichoplax adhaerens and Nematostella vectensis.
Eukaryotic proteins with divergent CPDadh domains
Several eukaryotic proteins were found to contain divergent CPDadh domains. They have different domain structures from proteins in the two groups as described earlier (see Fig. 2). A hypothetical protein from Chlamydomonas reinhardtii (gi|159466140) contains a transmembrane domain and several EF-hand calcium-binding domains. Divergent CPDadh domains are also present at the N-termini of several proteins containing NEK (NEver in mitosis Kinase) domain45 and RCC1 (regulation for chromosome condensation, with a β-propeller structure) domain46 in tunicata Ciona intestinalis (gi|198435791), insect Acyrthosiphon pisum (gi|193606321), and lancelet Branchiostoma floridae (gi|210108252, which also contains several tumor necrosis factor receptor (TNFR) domains47 at the C-terminus). One protein from marine choanoflagellate Monosiga brevicollis (gi|167516882) retains both the catalytic histidine and cysteine residues (see Fig. 1) and might still possess the peptidase activity. Interestingly, this protein lies in the middle of the bacterial group 1 proteins and eukaryotic group 1 proteins (Supporting Information Fig. 1), suggesting that the eukaryotic CPDadh domain could be acquired by horizontal gene transfer of a bacterial group 1 protein to an ancient eukaryotic organism.
Conclusions
We have identified a new cysteine peptidase family (CPDadh) homologous to the CPD domain of MARTX toxins. This new domain is present in a diverse collection of bacterial proteins, many of which are likely involved in cell adhesion. The substrates of the bacterial domains are unknown. One interesting possibility is that these proteins are autocleaved by the CPDadh domain, like the MARTX toxins. Eukaryotic members of CDPadh domains are found in cell surface protein NELF as well as some proteins co-occurring with signaling domains. Mutations in active site residues suggest that most of eukaryotic CPDadh domains are catalytically inactive. The functions of CPDadh domains in different groups of bacterial and eukaryotic proteins remain to be understood.
Materials and Methods
The PSI-BLAST program4 was used to search for homologs of the CPDmartx domain of V. cholerae (gi|153817921, range 3429–3637) against the NCBI nonredundant database (October 26, 2008; 7,124,886 sequences; 2,457,960,432 total letters), with an inclusion e-value cutoff of 0.001. Found homologs were clustered and representative sequences from each group were used to start new iterations of PSI-BLAST searches to ensure maximum coverage. Manual inspections of PSI-BLAST hits above the default e-value cutoff were conducted to find remote homologs of CPDmartx domains. The same PSI-BLAST search strategy was used for CPDadh domains. HHpred,10 a profile–profile based method, was used to find distant relationships for CPDadh and CPDmartx domains. The 3D-Jury Meta server11 was used for fold recognition for CPDadh domains. Domain architecture analysis was made by submitting sequences to domain database servers such as CDD,6 Pfam,7 and SMART,7 and by using the hmmpfam program of the HMMER package.7 Multiple sequence alignments were constructed by using the PROMALS3D program.19 Manual adjustment of the alignments was made with guidance from available 3D structure and secondary structure predictions made by PSIPRED.20 Sequence grouping and display of the groups were made by the CLANS program.21
Acknowledgements
The authors thank Rama Ranganathan, Aimee Shen, and Matt Bogyo for helping in discussions, and Lisa Kinch for critical reading of the manuscript and helpful suggestions. P.J.L. is a Damon Runyon Fellow, supported by the Damon Runyon Cancer Research Foundation. K.C.G. is supported by the Keck Foundation and Howard Hughes Medical Institute.




