Fast structural dynamics in reduced and oxidized cytochrome c
Abstract
The sub-nanosecond structural dynamics of reduced and oxidized cytochrome c were characterized. Dynamic properties of the protein backbone measured by amide 15N relaxation and side chains measured by the deuterium relaxation of methyl groups change little upon change in the redox state. These results imply that the solvent reorganization energy associated with electron transfer is small, consistent with previous theoretical analyses. The relative rigidity of both redox states also implies that dynamic relief of destructive electron transfer pathway interference is not operational in free cytochrome c.
Introduction
The influence of atomic scale structure on molecular recognition and catalysis continues to be intensively studied, but the role of structural dynamics is most often largely unknown.1 Protein-mediated electron transfer, integral to many critical biological processes, can be influenced by fast internal protein motions. Considerable effort has been directed at understanding the principles that govern through-protein electron transport,2, 3 but the exploration of how the nuclear and electronic structure of protein molecules influence electron tunneling remains a challenging and controversial problem.
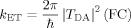
The present study was motivated by work that strongly suggests that dynamical effects on the TDA terms can be large.10, 11 Thus protein dynamics can modulate electron transfer by time-dependent variance of the energy gap or of the TDA terms. It is emphasized that the time scale of the fluctuations of the TDA terms most important for electron transfer is short, nanoseconds. This phenomenon is distinct from so-called gated electron transfer where a relatively slow larger scale conformational or orientational adjustment is required for electron transfer and is rate-limiting.12
Most investigations of the influence of sub-ns conformational fluctuations on electron transfer through modulation of the energy gap or TDA terms are based on molecular dynamics simulations. There has been little direct experimental investigation of protein dynamics in electron transfer proteins in this time regime, particularly in multiple redox states. Powerful nuclear magnetic resonance (NMR) relaxation methods now provide experimental access to protein dynamics over a wide range of time scales. Here we use NMR relaxation techniques13 to provide a site-resolved view of fast internal motions that are pertinent to the redox kinetics of cytochrome c.
Results and Discussion
Cytochrome c, the prototypical c-type cytochrome, is a component of the electron transport chain in mitochondria. We employed a H26N,H33N mutant which has superior expression properties but is otherwise largely indistinguishable from the wild-type horse protein.14 Resonance assignments of the diamagnetic reduced (spin zero) and paramagnetic oxidized (spin [1/2]) states were previously reported.15 Classical NMR relaxation provides access to the amplitude and time scale of motion through its influence on the spectral density defining observable relaxation parameters such as the spin-lattice relaxation time (T1).13 Here, nitrogen-15 relaxation was used to probe fast motion on the backbone and to characterize molecular tumbling. Deuterium relaxation was used to characterize the motion of methyl-bearing amino acid side chains. Relaxation data were interpreted using the Lipari-Szabo model free formalism.16 A recent re-evaluation of the model-free treatment reinforces confidence in its robustness with respect to highly asymmetric side chain motion.17
The squared generalized order parameters for amide NH bond vectors (O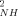 ) and for methyl group symmetry axes (O
) and for methyl group symmetry axes (O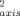 ) in reduced and oxidized cytochrome c are shown in Figure 1. As for many proteins.23 the reduced and oxidized cytochrome c O
) in reduced and oxidized cytochrome c are shown in Figure 1. As for many proteins.23 the reduced and oxidized cytochrome c O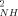 parameters for residues located in regular secondary structures are generally high. When sites having significant chemical exchange or paramagnetic relaxation contributions to the measured relaxation parameters are excluded, the average O
parameters for residues located in regular secondary structures are generally high. When sites having significant chemical exchange or paramagnetic relaxation contributions to the measured relaxation parameters are excluded, the average O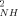 parameters for both the reduced (<O
parameters for both the reduced (<O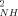 > ∼ 0.927) and oxidized (<O
> ∼ 0.927) and oxidized (<O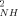 > ∼ 0.923) states point to an unusual rigidity for both forms of the protein. Also the average O
> ∼ 0.923) states point to an unusual rigidity for both forms of the protein. Also the average O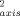 parameters for the reduced (<O
parameters for the reduced (<O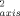 > ∼ 0.718) and the oxidized (<O
> ∼ 0.718) and the oxidized (<O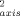 > ∼ 0.723) protein are also relatively high compared with most proteins.13 Both redox states of the protein are largely devoid of the so-called J-class of methyl sites undergoing sufficiently large excursions to result in rotamer interconversion.13 In keeping with previous results,24, 25 the present results suggest that the presence of large inflexible prosthetic groups suppresses the fast internal motion of proteins.
> ∼ 0.723) protein are also relatively high compared with most proteins.13 Both redox states of the protein are largely devoid of the so-called J-class of methyl sites undergoing sufficiently large excursions to result in rotamer interconversion.13 In keeping with previous results,24, 25 the present results suggest that the presence of large inflexible prosthetic groups suppresses the fast internal motion of proteins.
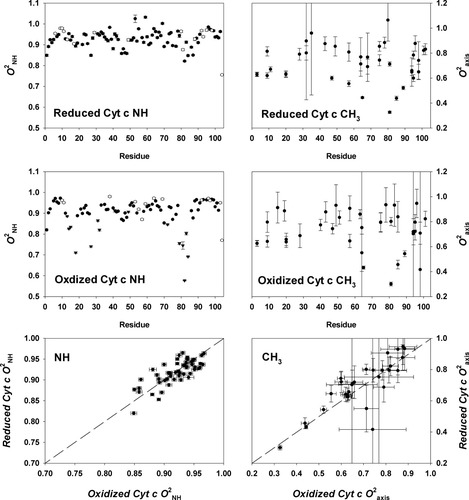
The amplitudes of ps-ns dynamics of the main chain NH (left panels) and of the methyl-bearing side chains (right panels) in reduced and oxidized [H26N,H33N]-horse cytochrome c at pH 5.8 and 20°C. Nitrogen-15 T1, T2 and NOE relaxation data were collected magnetic field strengths of 11.7 and 14.1 T essentially as described elsewhere18, 19. Deuterium T1 and T1ρ relaxation in methyl CH2D isotopomers was measured18-20 at magnetic fields of 11.7 and 14.1 Tesla using protein expressed during growth on 50% D2O. An effective 15N1H bond length of 1.04 Å, a 15N CSA tensor breadth of −170 ppm, and a deuterium quadrupolar coupling constant of 165 kHz were used in the calculations. Rotational anisotropy was determined to be negligible for both redox states. The isotropic form of the spectral density was therefore employed using the determined value for τm of 7.2 ns. Squared generalized order parameters and effective correlation times were obtained for individual sites using a grid-search algorithm.21 Precision was estimated using Monte Carlo sampling. O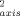 were calculated by dividing the obtained O2 by 0.111. The g-tensor of oxidized cytochrome c22 indicates a negligible paramagnetic contribution to deuterium relaxation and a significant (> 5%) contribution to amide 15N relaxation at only a few sites. Open circles correspond to sites with significant motion on the microsecond-millisecond timescale as detected by a R1R2 analysis. Solid triangles correspond to sites having a significant paramagnetic contribution (> 5%) to observed relaxation parameters. These sites are excluded from the redox state comparison (bottom panels). Lines of unit slope are drawn in the bottom panels.
were calculated by dividing the obtained O2 by 0.111. The g-tensor of oxidized cytochrome c22 indicates a negligible paramagnetic contribution to deuterium relaxation and a significant (> 5%) contribution to amide 15N relaxation at only a few sites. Open circles correspond to sites with significant motion on the microsecond-millisecond timescale as detected by a R1R2 analysis. Solid triangles correspond to sites having a significant paramagnetic contribution (> 5%) to observed relaxation parameters. These sites are excluded from the redox state comparison (bottom panels). Lines of unit slope are drawn in the bottom panels.
The O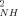 parameters of the reduced state are on average slightly higher than their counterparts in the oxidized state (<ΔO
parameters of the reduced state are on average slightly higher than their counterparts in the oxidized state (<ΔO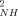 > ∼ 0.04; <|ΔO
> ∼ 0.04; <|ΔO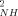 |> ∼ 0.05), as they are in yeast iso-1-cytochrome c.26 In slight contrast, the O
|> ∼ 0.05), as they are in yeast iso-1-cytochrome c.26 In slight contrast, the O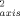 parameters of the oxidized state are on average higher than their counterparts in the reduced state (<ΔO
parameters of the oxidized state are on average higher than their counterparts in the reduced state (<ΔO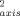 > ∼ 0.016; <|ΔO
> ∼ 0.016; <|ΔO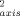 |> ∼ 0.046). On average, the oxidized state has slightly smaller amplitude of sub-ns motion than the reduced state. These results are generally in keeping with a vibrational analysis of the cytochrome c.27, 28 They may seem to contrast with the view provided by hydrogen exchange29 and NMR exchange broadening,30 but those probes sample much slower (infrequent) motions. Thus, it seems that the dynamical differences between the two redox states are largely restricted to slower time scales. A similar result has been observed using 15N-relaxation studies of backbone motion in Bacillus pasteurii cytochrome c, where the average O
|> ∼ 0.046). On average, the oxidized state has slightly smaller amplitude of sub-ns motion than the reduced state. These results are generally in keeping with a vibrational analysis of the cytochrome c.27, 28 They may seem to contrast with the view provided by hydrogen exchange29 and NMR exchange broadening,30 but those probes sample much slower (infrequent) motions. Thus, it seems that the dynamical differences between the two redox states are largely restricted to slower time scales. A similar result has been observed using 15N-relaxation studies of backbone motion in Bacillus pasteurii cytochrome c, where the average O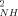 of the oxidized and reduced states were found to be essentially the same within experimental error. The results presented here also suggest that redox-dependent changes in residual protein entropy, expressed in the motions on the sub-ns time scale,1, 13, 17, 36 are small in cytochrome c. This contrasts with cyt b5 where significant changes in protein dynamics are apparent.31
of the oxidized and reduced states were found to be essentially the same within experimental error. The results presented here also suggest that redox-dependent changes in residual protein entropy, expressed in the motions on the sub-ns time scale,1, 13, 17, 36 are small in cytochrome c. This contrasts with cyt b5 where significant changes in protein dynamics are apparent.31
The fact that both redox states of cytochrome c are relatively rigid implies that a Pathways-type analysis based on a rigid structural framework is valid. Furthermore, the distributions of dynamics within the two states are closely similar. The near identity of the equilibrium structures of the two redox states32-34 suggests that the landscapes near the lowest energy structure of each redox state are also quite similar. These results indicate that the reorganization energy associated with electron self-exchange is small, consistent with a previous theoretical analysis.35 Finally, Balabin and Onuchic point out that electron transfer pathways need not be dynamically activated.11 Insofar as the relevant dynamics occur on the sub-ns time scale, the unusual rigidity of both redox states of cytochrome c implies that general dynamic relief of destructive interference of multiple pathways for electron transfer is not operational in free cytochrome c.
Abbreviations: FC, Franck-Condon density of states; O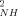 and O
and O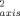 , Lipari-Szabo generalized order parameter for the amide NH bond vector and the methyl symmetry axis, respectively; TDA, electronic matrix element between the donor (D) and acceptor (A).
, Lipari-Szabo generalized order parameter for the amide NH bond vector and the methyl symmetry axis, respectively; TDA, electronic matrix element between the donor (D) and acceptor (A).
Methods
Nitrogen-15 T1, T2 and NOE relaxation data were collected magnetic field strengths of 11.7 and 14.1 T essentially as described elsewhere.18, 19 Deuterium T1 and T1ρ relaxation in methyl CH2D isotopomers was measured18-20 at magnetic fields of 11.7 and 14.1 Tesla using protein expressed during growth on 50% D2O. An effective 15N1H bond length of 1.04 Å, a 15N CSA tensor breadth of −170 ppm, and a deuterium quadrupolar coupling constant of 165 kHz were used in the calculations. Rotational anisotropy was determined to be negligible for both redox states. The isotropic form of the spectral density was therefore employed using the determined value for τm of 7.2 ns. Squared generalized order parameters and effective correlation times were obtained for individual sites using a grid-search algorithm.21 Precision was estimated using Monte Carlo sampling. O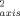 parameters were calculated by dividing the obtained O2 by 0.111. The g-tensor of oxidized cytochrome c22 indicates a negligible paramagnetic contribution to deuterium relaxation and a significant (>5%) contribution to amide 15N relaxation at only a few sites.
parameters were calculated by dividing the obtained O2 by 0.111. The g-tensor of oxidized cytochrome c22 indicates a negligible paramagnetic contribution to deuterium relaxation and a significant (>5%) contribution to amide 15N relaxation at only a few sites.




