Solution structure of the FCS zinc finger domain of the human polycomb group protein L(3)mbt-like 2
Abstract
Polycomb group proteins are epigenetic regulators that maintain patterns of gene expression over multiple rounds of cell division. Many of these proteins, including polyhomeotic and the MBT repeat containing proteins SCM and dSfmbt, contain an atypical C2C2 zinc finger with a characteristic phenylalanine–cysteine–serine sequence motif. The reoccurrence of this so-called FCS zinc finger in a variety of polycomb group proteins suggests that it has an important regulatory function. We have determined the solution structure of the FCS zinc finger of the human dSfmbt homologue L(3)mbt-like 2 (L3MBTL2). The structure consists of a β-hairpin followed by an α-helix. The zinc ligands are situated in the β-hairpin and at the N-terminus of the α-helix an arrangement typical of the treble clef class of zinc fingers. The structure is consistent with the proposal that FCS zinc fingers bind to regulatory RNAs.
Introduction
Polycomb group (PcG) proteins participate in the long-term epigenetic silencing of gene expression required for the acquisition and maintenance of cell fate.1 Deregulated expression of PcG proteins has been linked to malignant lymphomas, epithelial tumors, and osteosarcoma.2, 3 Most PcG proteins are components of large multiprotein complexes involved in the creation and recognition of chromatin modifications. The polycomb repressive complex 1 (PRC1) is an assembly of Drosophila PcG proteins that can bind to histone H3 methylated on lysine 27 and monoubiquitinate histone H2A. Two of the components of PRC1, polyhomeotic and Sex comb on midleg (SCM) contain an atypical C2C2 zinc finger domain with a characteristic phenylalanine–cysteine–serine sequence motif. This so-called FCS zinc finger (IPR012313) is also found in dSfmbt, a component of another PcG complex PhoRC.4 SCM and dSfmbt contain two and four copies, respectively, of the methyl lysine binding MBT repeat domain.5 In both SCM and dSfmbt, the FCS zinc finger is N-terminal while in polyhomeotic it is in the C-terminal region of the protein. FCS zinc fingers are present in the human homologues of polyhomeotic and dSfmbt, but are absent from human SCM like proteins. Human polyhomeotic homologues also form PRC1 like complexes while the dSfmbt homologue lethal (3) malignant brain tumor-like 2 (L3MBTL2) is a component of a complex that contributes to silencing of E2F- and Myc-responsive genes in quiescent cells.6 The function of the FCS zinc finger is unclear although it has been proposed that it may bind to noncoding RNAs.7 As part of an effort to learn more about the role of this domain, we have determined the solution structure of the FCS zinc finger of L3MBTL2.
Results and Discussion
The solution structure of residues 82–124 of L3MBTL2 was determined using standard nuclear magnetic resonance spectroscopic methods. The statistics for the 20 structures with the lowest overall energy are summarized in Table I. An overlay of the backbone atoms (Cα, C′, N) of these structures is shown in Figure 1(A). The structure consists of two β-strands (P87-C90 and I95-T98) that form an antiparallel sheet, followed by a 13-residue loop and a α-helix (V112-S120) [Fig. 1(B)]. The bound zinc ion is ligated by four cysteine residues. Two of them are at the end of the first β-strand (C90) and in the hairpin turn between the β-strands (C93). The third (C110) and the fourth (C114) cysteines are shortly before and at the beginning of the helix [Fig. 1(B)]. Other FCS zinc fingers have insertions and deletions within the loop [Fig. 2(A)] but are likely to have a very similar overall structure. The position of the zinc ligands is typical of the treble clef class of zinc fingers.9 A structure similarity search with VAST10 identified the Escherichia coli protein YacG,8 which binds and inhibits DNA gyrase,11 as the protein most closely related to the FCS zinc finger [Fig. 1(C)]. In both proteins the two pairs of zinc ligands are separated by a loop rather than elements of secondary structure, as is the case in most treble clef zinc fingers. With the exception of residues involved in maintaining the common fold there is little sequence conservation between these proteins and the structural similarity is unlikely to be functionally significant. The residues flanking the zinc binding cystine in the conserved FCS sequence also have a structural role. F109 is part of a hydrophobic core together with residues in the C-terminal end of the loop, the helix, and the β-hairpin. The side-chain of S111 forms a hydrogen bond to the backbone nitrogen of C114 and functions as the N-terminal cap of the helix. Two other conserved aromatic residues F102 and Y118 also form hydrophobic interactions that stabilize the fold. The phenylalanine helps to pack the loop onto the β-sheet and Y118, which is in the helix, interacts with residues both in the loop, including F109, and in the β-hairpin. A strictly conserved glycine residue that follows the second zinc ligand forms part of the β-hairpin. Three positions, two immediately preceding the FCS motif and one in the helix (residues K107, R108, and R116 in L3MBTL2) are occupied by positively charged residues in most FCS zinc fingers [Fig. 2(A)]. These residues are all on the same face of the molecule [Fig. 2(B)]. They do not appear to be involved in maintaining the structure of the domain and probably are functionally important. Treble clef zinc fingers are found in numerous proteins and have a variety of functions.9 In ribosomal protein L24E the treble clef zinc finger binds to 23S rRNA. This interaction is mediated primarily by hydrogen bond formation between the side chains of residues in the helix and the backbone of the RNA.12 As the potentially functionally important residues in the FCS zinc finger are on the same face of the fold that contacts rRNA in the ribosomal protein it is possible that it interacts with RNA in an analogous manner.
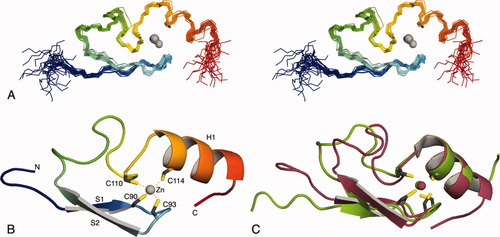
Solution structure of the FCS zinc finger domain of human L3MBTL2. (A) An overlay of the backbone atoms of the 20 lowest energy structures (PDB ID code 2w0t). The structures are colored blue to red from their N-terminus to the C-terminus. (B) Cartoon representation of the lowest energy structure. The secondary structure elements and the zinc coordinating residues are labeled. (C) Superimpositon of E. coli YacG (green; 1lv3; Ref.8) and the FCS zinc finger domain of L3MBTL2 (red). The backbones atoms (C′, N, Cα) of the secondary structure elements were used to superimpose the structures (YacG: T6-V17 and S29-G38; FCS: P87-T98 and V112-N121). Only the first 40 residues are shown for YacG.
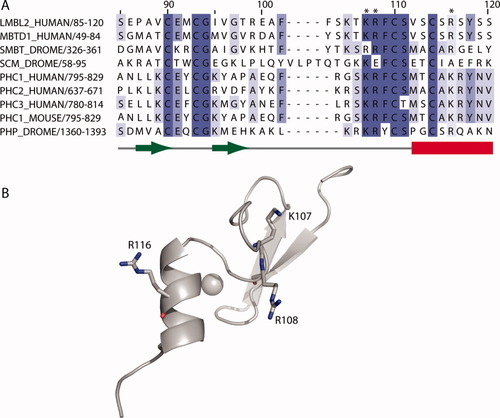
(A) Sequence alignment of FCS zinc finger domains from human, mouse, and Drosophila PcG proteins. The residues are colored according to their sequence identity. The conserved arginine and lysine residue are indicated. (B) Cartoon representation of the FCS zinc finger showing the positions of the conserved arginine and lysine residues.
| Structural constraints | |
| Intraresidue | 283 |
| Sequential | 170 |
| Medium-range (1 < |i – j| < 5) | 87 |
| Long-range (|i – j| ≥ 5) | 203 |
| Chi-1 angle constraints | 11 |
| TALOS constraints | 62 |
| Hydrogen bond constraints | 18 |
| Zinc coordination constraints | 10 |
| RDC constraints | 33 |
| Total | 877 |
| Statistics for accepted structures | |
| Statistics parameter (±SD) | |
| RMS deviation for distance constraints | 0.0085 ± 0.0027 |
| RMS deviation for dihedral constraints | 0.1391 ± 0.1147 |
| Mean CNS energy term (kcal mol−1 ± SD) | |
| E (overall) | 53.35 ± 20.60 |
| E (van der Waals) | 14.06 ± 5.10 |
| E (NOE and hydrogen bond constraints) | 4.55 ± 0.42 |
| E (chi-1 dihedral and TALOS constraints) | 0.54 ± 1.06 |
| RMS deviations from the ideal geometry (±SD) | |
| Bond lengths | 0.0017 ± 0.00038 |
| Bond angles | 0.3601 ± 0.0465 |
| Improper angles | 0.3331 ± 0.0833 |
| Average atomic RMSD from the mean structure (±SD) | |
| Residues 86–121 (N, Ca, C atoms) | 0.423 ± 0.1627 |
| Residues 86–121 (all heavy atoms) | 0.950 ± 0.1286 |
| Structural quality (residues 86–121) | |
| Residues in most favored regions of Ramachandran plot | 83.1% |
| Residues in additional allowed regions of Ramachandran plot | 14.7% |
| Residues in generously allowed regions of Ramachandran plot | 1.6% |
| Residues in disallowed regions of Ramachandran plot | 0.6% |
Although there is growing evidence that noncoding RNAs play an important role in PcG-mediated gene silencing,13 the identity of the RNAs involved and the mechanisms by which they act have not yet been fully established. Polyhomeotic binds to RNA and this activity depends upon its FCS zinc finger.7 This finding raises the possibility that FCS zinc fingers in general may link PcG proteins to regulatory RNAs. The data reported here is consistent with this proposal, however, given the wide range of functions carried out by structurally related proteins, the possibility that this domain has another role cannot be excluded. The availability of a structure for the FCS zinc finger will assist efforts to further define the role of this domain in the regulation of PcG function.
Materials and Methods
The DNA coding for residues 82–124 of human L(3)mbt-like 2 protein was amplified by PCR and cloned into a modified pRSETA (Invitrogen) expression vector. The resulting plasmid codes for fusion proteins with a N-terminal 6xhis-tag, the lipoyl domain of the Bacillus stearothermophilus dihydrolipoamide acetyltransferase and a TEV-cleavage site followed by the FCS zinc finger domain.
The resulting plasmid was transformed into Escherichia coli C41(DE3) cells. Cells were grown at 37°C to mid-log phase and induced with 1 mM IPTG. The temperature was reduced to 25°C and the cells were grown for a further 16 h. Cells were lysed by sonication, and the fusion protein was initially purified by Ni-NTA affinity chromatography. TEV protease digestion and a second affinity chromatography removed the lipoyl domain fusion tag. Final purification was performed with gel filtration using a HiLoad 26/60 Superdex 75 column (Amersham Pharmacia). NMR spectra were recorded at 25°C on Bruker DRX 500, DRX 600, and AVANCE 800 MHz spectrometers. The spectra were processed with TopSpin and analyzed with Sparky. Samples contained 1.0–1.3 mM protein in 10 mM MES (pH 6.3), 400 mM NaCl, and 5 mM 2-mercaptoethanole. Resonance assignments were obtained using standard methods and distance constraints were derived from NOESY spectra recorded with a mixing time of 125 ms. Hydrogen bond constraints were included for a number of backbone NH groups whose signals were observed in a 2D 1H-15N-HSQC recorded in D2O at 283 K. For hydrogen bond partners, two distance constraints were used where the distance (D)H-O(A) corresponded to 1.8–2.1 Å and (D)N-O(A) to 2.7–3.1 Å. Backbone torsional angle constraints were obtained from an analysis of C′, N, Cα Hα, and Cβ chemical shifts using the program TALOS.14 Residual dipolar coupling (RDC) values were measured in a 6.5% w/v acrylamide gel with higher buffer concentration (50 mM MES) to assure the correct pH in the gel.15 Control experiments under isomorphic conditions were performed in normal RDC buffer. The mean RDC values of two experiments were used for structure calculation. Structures were calculated using the standard torsion angle dynamics-simulated annealing protocol in the program CNS.16
Protein data bank deposition
Atomic coordinates have been deposited in the Protein Data Bank (accession code 2w0t).




