FRAGSITE2: A structure and fragment-based approach for virtual ligand screening
Review Editor: Nir Ben-Tal
Abstract
Protein function annotation and drug discovery often involve finding small molecule binders. In the early stages of drug discovery, virtual ligand screening (VLS) is frequently applied to identify possible hits before experimental testing. While our recent ligand homology modeling (LHM)-machine learning VLS method FRAGSITE outperformed approaches that combined traditional docking to generate protein–ligand poses and deep learning scoring functions to rank ligands, a more robust approach that could identify a more diverse set of binding ligands is needed. Here, we describe FRAGSITE2 that shows significant improvement on protein targets lacking known small molecule binders and no confident LHM identified template ligands when benchmarked on two commonly used VLS datasets: For both the DUD-E set and DEKOIS2.0 set and ligands having a Tanimoto coefficient (TC) < 0.7 to the template ligands, the 1% enrichment factor (EF1%) of FRAGSITE2 is significantly better than those for FINDSITEcomb2.0, an earlier LHM algorithm. For the DUD-E set, FRAGSITE2 also shows better ROC enrichment factor and AUPR (area under the precision-recall curve) than the deep learning DenseFS scoring function. Comparison with the RF-score-VS on the 76 target subset of DEKOIS2.0 and a TC < 0.99 to training DUD-E ligands, FRAGSITE2 has double the EF1%. Its boosted tree regression method provides for more robust performance than a deep learning multiple layer perceptron method. When compared with the pretrained language model for protein target features, FRAGSITE2 also shows much better performance. Thus, FRAGSITE2 is a promising approach that can discover novel hits for protein targets. FRAGSITE2's web service is freely available to academic users at http://sites.gatech.edu/cssb/FRAGSITE2.
1 INTRODUCTION
Virtual ligand screening (VLS) that uses computational tools to discover small molecules that might bind a protein target, not only has applications in drug discovery (Cases & Mestres, 2009; Kroemer, 2007; Reddy et al., 2007), but also in protein function analysis (Brylinski & Skolnick, 2010; Konc & Janežič, 2014). Traditionally, there are two broad categories of VLS methods: (a) structure-based docking methods use the high-resolution, three-dimensional (3D) structure of a protein target and dock ligand structures to the target protein (Allen et al., 2015; Jain, 2003; Kroemer, 2007; Trott & Olson, 2010); they then evaluate and rank the docked ligand structures using various scoring functions (Friesner et al., 2004; Kroemer, 2007; Ragoza et al., 2017; Wójcikowski et al., 2017). The advantage of docking methods is that they might discover novel binders to the target, whereas their disadvantage is that they are computationally expensive; (b) ligand-based methods use the known ligands of a given protein target to predict new binders based on the similarity to the physical–chemical properties of known ligands that bind the given protein target (Flower, 1998; Glen & Adams, 2006; Keiser et al., 2007; Willett, 2006). The advantage of ligand-based methods is that they are usually more accurate than docking, but they cannot discover novel small molecule binders that are chemically quite distinct from known binders, and they are not able to screen proteins with no known binders. To address these limitations, in our laboratory, we developed a series of ligand homology modeling (LHM)-based methods. Our suite of methods conceptually lies between the two traditional methods (Brylinski & Skolnick, 2008; Brylinski & Skolnick, 2009; Zhou et al., 2018, 2021; Zhou & Skolnick, 2013). They use lower resolution as well as high-resolution structures of protein targets to find similar binding pockets (template pockets) in the protein–ligand complex structures found in the Protein Data Bank (PDB; Bernstein et al., 1977); such pockets need not come from evolutionarily related proteins. They then use the corresponding ligands bound in the pockets of PDB structures as template ligands. To further expand the set of such template ligands, two additional databases: ChEMBL (Gaulton et al., 2012) and DrugBank (Wishart et al., 2006) for domain structure comparison (Zhou et al., 2018; Zhou & Skolnick, 2013) were employed. These template ligands are then used in a similar manner as ligand-based methods. Thus, the FINDSITE suite of methods has the advantage of having an accuracy comparable to ligand-based methods for those proteins having known binders, but importantly, they can be applied to proteins without known binders and are also computationally much less expensive than docking methods that not only require a high-resolution target structure, but also the 3D structures of the screened compounds.
The recent development of deep learning methods not only advanced the coverage of high-resolution structures of the human proteome and other species, for example, by AlphaFold (AlQuraishi, 2019; Jumper et al., 2021), but also improved the accuracy of scoring functions for docking methods (Ragoza et al., 2017; Wallach et al., 2015), for example, DenseFS is currently the best performing scoring function for VLS (Crampon et al., 2022; Imrie et al., 2018; Singh et al., 2023). Other algorithms, like the latest version of ConPLex (Singh et al., 2023), are trained and tested on databases of binding affinity or pairwise classification involving many proteins. Scoring functions trained on protein–ligand pairwise affinities (e.g., BindingDB; Liu et al., 2007) or classification (e.g., BIOSNAP: https://snap.stanford.edu/biodata/), do not work for VLS. For example, the RF-score trained on the BindingDB affinity dataset has to be retrained on the DUD-E set (Wójcikowski et al., 2017); that is, they are not transferable to protein families lacking any known small molecule binders. The latest generative language model of ConPLex trained on BIOSNAP—a protein–drug interaction network database also has to include the decoys of DUD-E set to train their model to better classify the actives from decoys of the testing DUD-E subset (Singh et al., 2023). Without the inclusion of decoys from the protein target of interest in their training, RF-score or ConPLex performs poorly in discriminating actives/drugs from decoys for a given protein target (Singh et al., 2023; Wójcikowski et al., 2017).
However, how good ConPLex is for VLS compared with other VLS scoring functions is unknown as it does not report the results of conventional measures such as the enrichment factor, AUPR (area under the precision-recall curve) or AUROC (area under the ROC curve). The new scoring function DenseFS (Imrie et al., 2018) coupled with traditional AutoDock Vina (Trott & Olson, 2010) and trained on the DUD-E set (Mysinger et al., 2012) has close overall performance for the DUD-E set as FRAGSITE when the sequence cutoff is set to 80% between training and testing proteins. It has an AUROC and ROC 1% enrichment factor (ROCEF1%: the enrichment at a 1% false positive rate, which is a different metric from the enrichment factor of the top 1% ranked molecules) of 0.92 and 48.0, respectively, whereas FRAGSITE (Zhou et al., 2021) has scores of 0.91 and 61.5. However, the performance of a DenseFS trained family-specific model and its prediction for unrelated or remote family proteins is uncertain. In practice, DenseFS is limited to proteins belonging to the four families in its training set. Furthermore, most docking methods rely on predefined binding pockets to sample reliable binding poses (Allen et al., 2015; Trott & Olson, 2010). Similar to family-specific training, this limits the protein coverage of the method since the majority of proteins do not have the structures of protein–ligand complexes. Another concern with deep learning on docked poses is that errors generated by docking pass to the scoring function models (unlike AlphaFold for protein structure prediction that learns from experimental structures (AlQuraishi, 2019; Jumper et al., 2021)). The trained scoring function could rank wrongly posed ligands higher. When the same ligand is posed correctly by more accurate docking methods or by different random generators, the scoring function could incorrectly rank the ligand. Furthermore, the computational expense might preclude large-scale applications of docking-related scoring functions. In contrast, the FINDSITE and FRAGSITE suite of methods cover 97% of human protein sequences, with similar coverage for other species, and all can be set up as web servers (Zhou et al., 2018, 2021; Zhou & Skolnick, 2013).
Although FRAGSITE (Zhou et al., 2021) outperforms the state-of-the-art deep learning based scoring function approaches, e.g. DenseFS (Imrie et al., 2018) in terms of ROC enrichment (ROCEF1%: 61.5 vs. 48.0), the main drawback of the current FINDSITE suite of methods, including FRAGSITE, is that since they all used template ligands to search for new binders, they are not able to discover novel binders that are chemically very dissimilar from the template ligands. To overcome this disadvantage, we utilize the information from the template pockets rather than the template ligands to develop FRAGSITE2. FRAGSITE2 is more closely related to structure-based methods, but without the need for high-resolution target structures, 3D structures of the ligands, and docked ligand poses. By constructing feature vectors using pocket information in combination with screened ligands, we apply a boosted regression tree machine learning method as in FRAGSITE (Zhou et al., 2021) to train the model on the DUD-E set (Mysinger et al., 2012) and do a modified leave one out cross-validation (LOOCV) test with a sequence cutoff of 80% (the training protein targets have sequence identity <80% to the given testing protein target) to fairly compare to other state-of-the-art deep learning methods such as the DenseFS function (Imrie et al., 2018) and RF-score-VS (Wójcikowski et al., 2017). An independent test on the DEKOIS2.0 set (Bauer et al., 2013) was also performed; we note here that FRAGSITE2 was not retrained for the DEKOIS2.0 set. In what follows, in addition to comparison to the state-of-the-art VLS methods FINDSITEcomb2.0 and FRAGSITE, we also compare the performance of FRAGSITE2 to the state-of-the-art deep learning-based DenseFS scoring function (Imrie et al., 2018) on the DUD-E set and a typical machine learning scoring function, RF-score-VS (Wójcikowski et al., 2017), for VLS on the DEKOIS2.0 set. As to the latest generative language model of ConPLex (Singh et al., 2023), since it was only tested on a 31 target subset of the DUD-E set that was split from the 57 targets classified into 4 protein families and evaluated only for classification (separating actives from decoys) it does not report the results of conventional measures such as the enrichment factor, AUPR or AUROC; thus, we cannot compare FRAGSITE2 with ConPLex (Singh et al., 2023).
2 RESULTS
2.1 Benchmarking on the DUD-E set
The DUD-E set (Mysinger et al., 2012) is commonly used by VLS methods for benchmarking and training of machine learning scoring functions (Crampon et al., 2022). Although some of the machine learning scoring functions have been trained on pairwise protein–ligand binding affinity or classification (Crampon et al., 2022), they are usually not good for VLS as demonstrated by the RF-score-VS study of Wójcikowski et al. (2017). FRAGSITE2 also uses the DUD-E set for training and cross-validation testing. Here, cross-validation is performed by removing proteins having an amino sequence identity >80% to the given testing target from training. This is similar to the so-called “vertical split” scenario of RF-score-VS in Ref. (Wójcikowski et al., 2017). To test the ability of a method to discover novel actives with respect to template ligands (here, in benchmarking tests, template ligands are from homologous proteins having a sequence identity less than 80% to the given testing target; Zhou et al., 2018), we evaluate subsets of actives having a Tanimoto coefficient (TC) less than cutoff as well as with no cutoff.
Table 1 summarizes FRAGSITE2's results in comparison to FINDSITEcomb2.0 and FRAGSITE. In practice, one can combine all three methods using the maximal precision for each screened ligand; we call this combined method, FRAGSITEcomb in Table 1. We note that DUD-E set has a decoy/active ratio of around 60 resulting in an EF1% ~ 60 for the case of perfect ranking. Overall, without a TC cutoff to template ligands, the ligand-based methods FINDSITEcomb2.0 (EF1% = 37.20) and FRAGSITE (EF1% = 41.44) perform better than structure-based FRAGSITE2 (EF1% = 32.72) in terms of enrichment at the top 1% ranked list, whereas the combined approach FRAGSITEcomb has the best EF1% of 41.69. However, FRAGSITE2 still performs better than FINDSITEcomb2.0 and FRAGSITE for the number of targets having EF1% > 1 (better than random selection). With no cutoff, FRAGSITE2 has 99 targets with an EF1% > 1, whereas FINDSITEcomb2.0 and FRAGSITE each have 95 and 97 targets, respectively. This trend continues across all TC cutoffs. Most notably when the TC ≤ 0.8, FRAGSITE2 has a better mean EF1% of 37.06 compared with 30.89 of FRAGSITE and 17.08 of FINDSITEcomb2.0. Note that the performance of FRAGSITE2 is much less sensitive to which set of ligands are included which is in contrast to the performance of both FRAGSITE and FINDSITEcomb2.0. This indicates that FRAGSITE2's performance is independent of the actives' similarity to template ligands, a characteristic well suited for discovering novel actives (see Figure 1). The combined approach, FRAGSITEcomb, is the best performing method when TC ≥ 0.8. Compared with the previous typical machine learning score, RF-score-VS, with a mean EF1% ~ 12.5 in a vertical split scenario on the DUD-E set (Wójcikowski et al., 2017), FRAGSITE2's EF1% = 32.72 is significantly better (see Table 1, no cutoff).
| FINDSITEcomb2.0 | FRAGSITE | FRAGSITE2 | FRAGSITEcomba | |||||
|---|---|---|---|---|---|---|---|---|
| TC cutoff to template ligands | EF1% | No. of EF1% > 1 | EF1% | No. of EF1% > 1 | EF1% | No. of EF1% > 1 | EF1% | No. of EF1% > 1 |
| None | 37.20 | 95 | 41.45 | 97 | 32.72 | 99 | 41.69 | 100 |
| 0.9 | 30.46 | 93 | 42.22 | 96 | 38.75 | 97 | 45.43 | 99 |
| 0.8 | 17.08 | 74 | 30.89 | 92 | 37.06 | 94 | 37.67 | 96 |
| 0.7 | 8.90 | 39 | 21.58 | 69 | 33.36 | 82 | 31.11 | 82 |
| 0.6 | 4.74 | 13 | 13.47 | 32 | 32.87 | 58 | 28.21 | 51 |
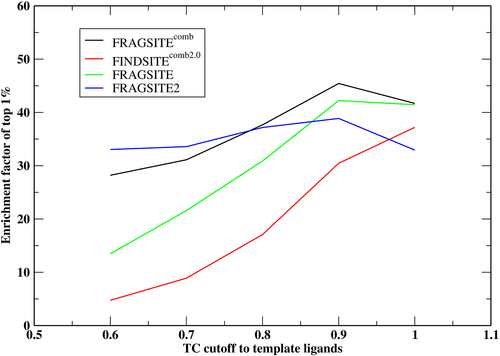
Next, we analyze the diversity of actives within the top 1% of the ranked order list. The mean number of clusters of each method is given in Table 2. Except for the case when no TC cutoff to template ligands is used, FRAGSITE2 has a mean number of clusters greater than those of FRAGSITE. The composite approach, FRAGSITEcomb, performs best when TC ≥ 0.8. This indicates that the higher EF1% of FRAGSITE than FRAGSITE2 when no TC cutoff is applied (41.44 vs. 32.72) is due to more similar actives within the top-ranked list. In terms of the diversity of actives, they are very close (31.60 vs. 29.19). Thus, FRAGSITE2 is better than FRAGSITE in discovering diverse actives.
| TC cutoff to template ligands | FINDSITEcomb2.0 | FRAGSITE | FRAGSITE2 | FRAGSITEcomb |
|---|---|---|---|---|
| None | 27.98 | 31.60 | 29.19 | 34.64 |
| 0.9 | 15.33 | 21.97 | 23.60 | 26.60 |
| 0.8 | 7.42 | 12.52 | 16.77 | 17.05 |
| 0.7 | 2.66 | 6.45 | 11.19 | 10.83 |
| 0.6 | 0.26 | 1.67 | 5.02 | 4.75 |
- Note: TC = 0.8 cutoff was used in clustering. Bold numbers are the best performing results.
- Abbreviation: TC, Tanimoto coefficient.
We next compared FRAGSITE2 to the state-of-the-art deep learning based method DenseFS scoring function (Imrie et al., 2018) using the AUPR (Davis & Goadrich, 2006) as the relevant metric. The results for mean AUPRs are given in Table 3. Notably, ligand homology-based methods FINDSITEcomb2.0 and FRAGSITE are the best performing single (noncombined) methods with AUPRs of 0.508 and 0.591, respectively, compared with DenseU's (a version of DenseFS without family-specific training) 0.368 and DenseFS's 0.443. FRAGSITE2's 0.465 is better than those of DenseU's and DenseFS's. FRAGSITE2's ROCEF1% = 49.11 is slightly better than DenseFS's 47.99. Thus, as a structure-based approach, FRAGSITE2 outperforms the structure-based, deep learning DenseU and DenseFS scoring functions. The combined approach, FRAGSITEcomb, with an overall AUPR = 0.606 and an ROCEF1% = 63.84, has superior performance to the best performing single method.
| AUPR | ROCEF1% | |
|---|---|---|
| DenseUa | 0.368 | 40.92 |
| DenseFS | 0.443 | 47.99 |
| FINDSITEcomb2.0 | 0.508 | 52.67 |
| FRAGSITE | 0.591 | 61.54 |
| FRAGSITE2 | 0.465 | 49.11 |
| FRAGSITEcomb | 0.606 | 63.84 |
- Note: Bold numbers indicate best performing method.
- Abbreviations: AUPR, area under the precision-recall curve; ROCEF1%, ROC 1% enrichment factor.
- a DenseFS scoring function without family-specific training (Imrie et al., 2018). DenseU and DenseFS results are from Tables S7and S9 of Ref. (Imrie et al., 2018).
2.2 Testing on the DEKOIS2.0 dataset
The DEKOIS2.0 dataset (Bauer et al., 2013) was used to benchmark FRAGSITE2 as an independent testing dataset from the training DUD-E set. DEKOIS2.0 is based on BindDB bioactivity data (Livyatan et al., 2015) and provides 81 structurally diverse benchmark sets for a wide variety of different target classes. It has a decoy/active ratio of 30; meaning the maximal possible EF1% is 30, which is smaller than 60 found for the DUD-E set. To compare to other previous methods, we again use an 80% sequence identity cutoff between testing the DEKOIS2.0 targets and the training DUD-E targets. Table 4 summarizes the performance of FRAGSITE2 on the DEKOIS2.0 set in comparison to FINDSITEcomb2.0 and FRAGSITE, and the combined approach, FRAGSITEcomb. Figure 2 shows the dependence of EF1% on the TC cutoff on the DEKOIS2.0 set. Both Table 4 and Figure 2 show a similar trend in results as was seen for the DUD-E set. With no TC cutoff to template ligands, FRAGSITE2 has a close EF1% (15.87) to that of FRAGSITE (17.72), and the numbers of targets with an EF1% > 1 are also close (70 vs. 72). The combined approach, FRAGSITEcomb, has a slightly better EF1% (18.30) compared with all three individual methods and has a larger number of targets, 75, with an EF1% > 1. With a TC cutoff = 0.7, both FRAGSITE2's EF1% and the number of targets with EF1% > 1 are significantly better than those of FRAGSITE's (23.63 vs. 11.66, 57 vs. 41, respectively).
| FINDSITEcomb2.0 | FRAGSITE | FRAGSITE2 | FRAGSITEcomb | |||||
|---|---|---|---|---|---|---|---|---|
| TC cutoff to template ligands | EF1% | No. of EF1% > 1 | EF1% | No. of EF1% > 1 | EF1% | No. of EF1% > 1 | EF1% | No. of EF1% > 1 |
| None | 16.67 | 66 | 17.72 | 72 | 15.87 | 70 | 18.30 | 75 |
| 0.9 | 13.49 | 58 | 19.17 | 70 | 18.58 | 67 | 19.95 | 70 |
| 0.8 | 7.02 | 42 | 15.02 | 58 | 19.67 | 66 | 18.95 | 63 |
| 0.7 | 3.13 | 13 | 11.66 | 41 | 23.63 | 57 | 21.97 | 54 |
| 0.6 | 0.52 | 3 | 10.02 | 17 | 18.55 | 30 | 15.74 | 28 |
- Note: Bold numbers are the best performing results.
- Abbreviations: EF1%, 1% enrichment factor; TC, Tanimoto coefficient.
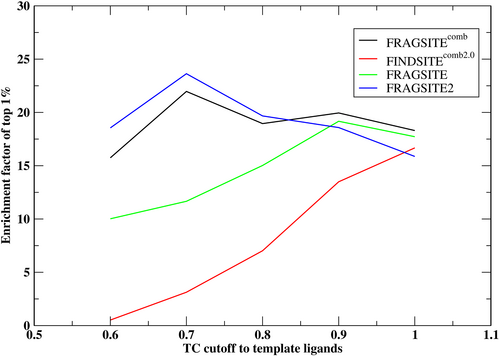
We note that as the TC cutoff becomes smaller, FRAGSITE2's mean EF1% increases. This is due to combination of three effects: (a) a possible performance change for ligands more dissimilar to template ligands, which is small for FRAGSITE2 because it does not sense this information, but it could carry on information from the training set that ligands dissimilar to template ligands might be undertrained. This could be the reason that the number of targets having EF1% > 1 decreases; (b) increase in the value of EF1% when a smaller subset of actives are evaluated (the decoy/active ratio increase results in a larger upper bound); (c) for a TC cutoff ≤0.7, some targets have no actives, thus, are not included in evaluation. The mean numbers of active clusters within the top 1% ranked list are given in Table 5. FRAGSITE2 has the best performance when the TC cutoffs to template ligands are ≤0.8, whereas the combined approach is best for a TC = 0.9 cutoff and no cutoff. Overall, FRAGSITE2's performance is better than FRAGSITE and FINDSITEcomb2.0 in terms of the diversity of top-ranked actives.
| TC cutoff to template ligands | FINDSITEcomb2.0 | FRAGSITE | FRAGSITE2 | FRAGSITEcomb |
|---|---|---|---|---|
| None | 4.46 | 5.09 | 4.68 | 5.25 |
| 0.9 | 2.25 | 3.95 | 4.01 | 4.25 |
| 0.8 | 0.98 | 2.85 | 3.46 | 3.37 |
| 0.7 | 0.22 | 1.76 | 2.47 | 2.30 |
| 0.6 | 0.0 | 0.67 | 0.99 | 0.91 |
- Note: TC = 0.8 cutoff was used in clustering. Bold numbers are the best performing results.
- Abbreviation: TC, Tanimoto coefficient.
To compare to the RF-score-VS scoring function (Wójcikowski et al., 2017) on the same DEKOIS2.0 76 target subset, we also excluded four overlapping structures between DUD-E and DEKOIS 2.0: A2A: “2p54,” HDAC2: “3l3m,” PARP-1: “3eml,” PPARA: “3max,” and SIRT2 having no crystallized ligand. (For FRAGSITE2, the overlapped targets were naturally excluded by using the 80% sequence identity cutoff between training and testing targets, as it does not require any experimental binders of the target protein). Furthermore, the evaluation was performed only on actives and decoys having a TC < 0.99 to any ligand/decoy in the training DUD-E set. For this subset of targets and ligands, the performance of the RF-Score-VS v2 and v3 were EF1% = 9.84 and EF1% = 7.81, respectively (Wójcikowski et al., 2017), whereas FRAGSITE2 achieves an EF1% = 20.18, more than double that of the best RF-score-VS.
The above two testing results indicate that for actives close to template ligands (or known ligands of the given target), FRAGSITE2 performs comparably to the state-of-the-art FRAGSITE which outperforms the recent best performing family based deep learning scoring function, DenseFS (Imrie et al., 2018). FRAGSITE2 doubled the early (1%) enrichment factor of conventional machine learning scoring function RF-score-VS. For actives remote to template ligands/known ligands, FRAGSITE2 obviously performs better than FRAGSITE and FINDSITEcomb2.0. In a practical scenario, the combined approach that simply takes the maximal predicted precision of each screened molecule from the three methods (FINDSITEcomb2.0, FRAGSITE, FRAGSITE2) has a performance close to that of the best possible performing algorithm when TC > 0.8, otherwise, FRAGSITE2 alone should be used.
2.3 Comparison to alternative learning method and protein features
Here, we examine an alternative learning method to boosted tree regression and other protein features to tease out the strengths/weaknesses of FRAGSITE2. The deep neural network method of multiple layer perceptron (MLP) regression MLPRegressor implemented in the Scikit-learn kit (Pedregosa et al., 2011) was applied on the same features as in FRAGSITE2 for the DUD-E set. After empirically optimizing a number of hidden layers and a number of hidden layer neurons, activation function, and maximal number of iterations (max_iter, others are default), we found one hidden layer with 800 neurons, tanh activation function, max_iter = 5000 gave the best results of mean (EF1% = 33.77, ROCEF1% = 51.75, AUPR = 0.464). They are very close to those of FRAGSITE2's: (32.72, 49.11, 0.465). Increasing max_iter to 5500 will result in over training and slightly decreasing in performance to (32.78, 50.98, 0.452). Adding an additional hidden layer, for example, a layer with 400 neurons, will also decrease performance to (32.24, 49.77, 0.436). Even though for the DUD-E set, MLP gives ~3% better EF1% than FRAGSITE2 does, it has worse EF1% performance for the 81 target DEKOIS2.0 set (13.46 vs. 15.87). Thus, FRAGSITE2's better performance than deep learning-based methods (e.g., DenseFS for DUD-E set) is due to its better features, not its learning method. In addition, boosted tree regression is more robust than a deep network on independent test set such as the DEKOIS2.0 set.
To further demonstrate our better pocket-based protein features, we tested alternative features for protein targets of the DUD-E set from the pretrained protein language model by deep learning (Elnaggar et al., 2022) as were used in the ConPlex work (Singh et al., 2023) for protein targets. The 1024 dimension embeddings of the ProtBert model (used by ConPlex and shown to be the best embedding for protein–ligand binding prediction in that work) were used to replace the 20 dimension features of FRAGSITE2's pocket-derived features for each protein target of the DUD-E set. Exactly the same training procedure as in FRAGSITE2 was applied to these 1905 dimension features. This resulted in mean (EF1% = 28.04, ROCEF1% = 40.66, AUPR = 0.387) that are much worse than those of FRAGSITE2's. Thus, FRAGSITE2's protein target features are much better than if a deep learning pretrained protein language model for protein–ligand/drug binding prediction is used. This might be due to the fact that our pocket-based features capture more specific ligand binding features than the language embedding of whole protein sequence.
3 DISCUSSION
Our results show that on the DUD-E set, for ligands having a TC < 0.7 to template ligands, the 1% enrichment factor (EF1%) of FRAGSITE2 is 33.4 compared with 21.6 of FRAGSITE and 8.9 of FINDSITEcomb2.0. On the 81 target DEKOIS2.0 set with the same TC < 0.7 to template ligands, FRAGSITE2 has an EF1% of 19.1, FRAGSITE has an EF1% of 11.7 while FINDSITEcomb2.0 has an EF1% of 3.1. Compared with the deep learning DenseFS scoring function (Imrie et al., 2018) with an AUPR of 0.443, FRAGSITE2 has a better AUPR of 0.465 and ROCEF1% of 49.1 versus 48.0 for the DUD-E set. Compared with the RF-score-VS on a 76 target subset of DEKOIS2.0 and actives and decoys with a TC < 0.99 to training DUD-E ligands, FRAGSITE2 has an EF1% of 20.2, whereas RF-score-VS has an EF1% of 9.8. Thus, FRAGSITE2 outperformed state-of-the-art VLS methods, in particular for finding novel binders that are chemically dissimilar (TC < 0.7) from existing binders (template ligands) of a given target's homologous proteins (or/and self). We further note that the FRAGSITE2 web service is freely accessible to academic users at http://sites.gatech.edu/cssb/FRAGSITE2.
Although the ligand homology-based methods FINDSITEcomb2.0 and FRAGSITE, like other ligand-based methods, in general, perform better than traditional structure-based methods, they suffer from the limitation that they tend to discover ligands that are similar to existing ones as demonstrated in Tables 2 and 5. At all TC cutoff <1 values, the numbers of active clusters within the top 1% ranked molecules are not better than those of the structure-based FRAGSITE2 method. Instead, when actives are chemically dissimilar from existing ligands, structure-based FRAGSITE2 shows better performance as assessed by ligand diversity. In practice, one can combine the two kinds of methods using the predicted precision of each molecule in FRAGSITEcomb, which has better performance in the TC > 0.8 range. Since many state-of-the-art deep learning methods, for example, ConPLex (Singh et al., 2023), are not fully benchmarked on conventional VLS sets, FRAGSITE2 is not able to compare to them. However, compared with the DenseFS function (Imrie et al., 2018), FRAGSITE2 is better in terms of ranking actives at the top as demonstrated by the AUPR measure (0.465 vs. 0.443). We showed that FRAGSITE2's better performance is due to its better protein target features derived from pockets. Another advantage of FRAGSITE2 compared with other docking and deep learning-based methods is that its computational cost is much less expensive. It can be easily run on a single desktop or laptop computer or readily implemented as web server.
The reason that FRAGSITE2 has superior performance independent of the actives' structure closeness to known/homologous protein binders is that the training and prediction features have not used the information from known binding ligands. Instead, it uses only the template pocket composition profile that encodes the structure and sequence information of the target's binding sites. Possible future improvements of FRAGSITE2 could involve a better representation of target pocket/binding sites beyond using their amino acid composition. Finally, a possibly better description of ligand fragments by decomposing the ligand into conservative fragments that bind to a specific pocket conformation is also currently under investigation.
4 MATERIALS AND METHODS
The flowchart of FRAGSITE2 is shown in Figure 3. FRAGSITE2 employs the FINDSITEfilt component of FINDSITEcomb2.0 (Zhou et al., 2018) that only uses a PDB protein–ligand complex for binding site prediction and for deriving template ligands. In FINDSITEcomb2.0, a template protein must have a TM-score (Zhang & Skolnick, 2004) greater than 0.6 to the target protein's structure and at least 80% of the template sequence must be aligned to the target sequence. A sequence cutoff is applied in benchmarking mode to exclude templates whose sequence identity > the cutoff for selecting template pockets. Then, template pockets are selected using up to the top 75 pockets from the PDB ligand–protein complex structures (Zhou et al., 2018).
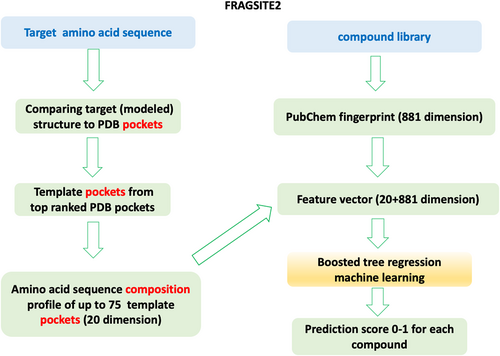
4.1 Training and testing datasets
We used the DUD-E (Mysinger et al., 2012) ligand virtual screening benchmark dataset for both training and testing. We conducted a modified LOOCV by excluding all targets having a sequence identity >80% to the given tested protein target. We used the DUD-E (Mysinger et al., 2012) ligand virtual screening benchmark dataset for both training and testing. We conducted a modified LOOCV by excluding all targets having a sequence identity >80% to the given tested protein target. This 80% identity cutoff is used only for template pocket selection and training target inclusion for given testing target for fair comparison to other state-of-the-art methods such as the DenseFS function (Imrie et al., 2018) and RF-score-VS (Wójcikowski et al., 2017) that used this cutoff to separate training and testing targets. However, for protein target structure modeling, we applied a 30% sequence cutoff to templates. In training the boosting tree function (1) for ligand–protein binding, the objective function value is assigned as: 1 if the molecule is a true binder of the target (in the DUD-E benchmarking set; Mysinger et al., 2012, the active ligands), and 0 if the molecule is not a binder (decoys in DUD-E). Since overall, DUD-E has an active to decoy ratio around 0.016, we randomly picked ~10% decoys and used all actives in training resulting in around 160,000 protein–ligand pairs.
To avoid any bias in the DUD-E set that favors a machine learning method with training and testing on the same set, we also tested FRAGSITE2 on an independent set from the training DUD-E set, DEKOIS2.0 (Bauer et al., 2013). This set has 81 structurally diverse targets with an actives to decoys ratio around 0.033 and is based on BindDB bioactivity data (Livyatan et al., 2015). To compare to previous work, we use an 80% instead of 30% sequence identity cutoff between testing targets and training DUD-E targets. We note that there are some other VLS benchmarking sets, for example, MUV (Rohrer & Baumann, 2009) and LIT-PCBA (Tran-Nguyen et al., 2020). However, since the MUV set has a total of only 17 targets and LIT-PCBA has 15 targets, they represent a very small number of protein families, and the small number of targets make them statistically insufficient to distinguish between methods. The performance of a few outliers could dominate the overall performance.
4.2 Assessment
To compare to the DenseFS score (Imrie et al., 2018), for a cutoff independent evaluation, we prefer AUPR, the area under the precision-recall curve (Davis & Goadrich, 2006) to AUC (area under the ROC curve) and the ROC 1% enrichment factor (ROCEF1%: enrichment at 1% false positive rate) that is slightly different from the above EF1%. AUPR is a better measure than AUC to distinguish the ability of methods to rank positives in the very top ranks when true positives are rare and only the very top-ranked ones are tested as is the case in VLS (Davis & Goadrich, 2006).
The precision score is useful for judging if the prediction is confident or not. To derive the predicted precision, we merge all the LOOCV predictions for actives and decoys of all targets from the DUD-E dataset (Mysinger et al., 2012) and bin the score Sfrg from 0 to 1 using . The precision score is used for the combined approach, FRAGSITEcomb, that simply takes the maximal precision score from the three methods: FINDSITEcomb2.0, FRAGSITE, and FRAGSITE2 to select a given screened molecule.
To test the ability of methods to discover novel binders from existing binders of self or close template proteins, we also evaluate for those actives that have a TC less than a cutoff to the template ligands. All TC are calculated by Open Babel (O'Boyle et al., 2011) with the FP2 option which indexes small molecule fragments based on linear segments of up to seven atoms (somewhat similar to the Daylight fingerprints; Anonymous, 2007).
AUTHOR CONTRIBUTIONS
Hongyi Zhou: Conceptualization; methodology; writing – original draft; formal analysis; data curation; validation. Jeffrey Skolnick: Conceptualization; funding acquisition; writing – review and editing; supervision.
ACKNOWLEDGMENTS
We thank Bartosz Ilkowski for computing support and Jessica Forness for proof-reading the article. This project was funded by R35GM118039 of the Division of General Medical Sciences of the NIH.




