Controlling the interaction between CaMKII and Calmodulin with a photocrosslinking unnatural amino acid
Abstract
Using unnatural amino acid mutagenesis, we made a mutant of CaMKII that forms a covalent linkage to Calmodulin upon illumination by UV light. Like wild-type CaMKII, the L308BzF mutant stoichiometrically binds to Calmodulin, in a calcium-dependent manner. Using this construct, we demonstrate that Calmodulin binding to CaMKII, even under these stochiometric conditions, does not perturb the CaMKII oligomeric state. Furthermore, we were able to achieve activation of CaMKII L308BzF by UV-induced binding of Calmodulin, which, once established, is further insensitive to calcium depletion. In addition to the canonical auto-inhibitory role of the regulatory segment, inter-subunit crosslinking in the absence of CaM indicates that kinase domains and regulatory segments are substantially mobile in basal conditions. Characterization of CaMKIIL308BzF in vitro, and its expression in mammalian cells, suggests it could be a promising candidate for control of CaMKII activity in mammalian cells with light.
1 INTRODUCTION
Many ion channels, enzymes, and signaling proteins connect to calcium signaling by binding the small calcium-binding protein Calmodulin. Calcium binds to Calmodulin at four sites, allowing it to expose hydrophobic pockets on its paired EF-hand motifs, and envelop binding motifs on its target proteins (Chin & Means, 2000; Jurado et al., 2022). One of Calmodulin's best-studied targets is Ca2+-Calmodulin-dependent protein kinase II (CaMKII; Herring & Nicoll, 2016; Nicoll, 2017; Incontro et al., 2018).
CaMKII isozymes () regulate a plethora of cellular processes, including myocardial signaling and neuronal plasticity involved in learning and memory (Takemoto-Kimura et al., 2017). A unique feature of this kinase is that it is organized as a dodecameric holoenzyme, made out of 12 subunit monomers. Each monomer of CaMKII comprises an N-terminal Serine/Threonine kinase domain, followed by a regulatory domain, a linker region, and an association or “hub” domain (Figure 1a), which mediates self-oligomerization (Chao et al., 2011; Myers et al., 2017). In basal conditions, CaMKII is thought to be inhibited through stable docking of its regulatory domain to the substrate binding site. Once Ca2+ enters the cell and binds to Calmodulin, Ca2+-calmodulin targets the regulatory domain of CaMKII and activates it by promoting an order-to-disorder transition of the inhibitory helix in the regulatory domain, which in turn frees the substrate binding site (Rellos et al., 2010). Subsequent trans-autophosphorylation at residue T286 (CaMKII numbering), in the regulatory domain, adjacent to the Ca2+-calmodulin binding site, is enough to prevent rebinding of the regulatory domain to the substrate binding site. In principle and according to consensus models, CaMKII no longer requires Ca2+-calmodulin for its activity, and it remains active even after the initial Ca2+ stimulus has returned to baseline. In other words, calcium activation converts CaMKII from a Ca2+-triggered kinase into a constitutively-active enzyme. This bistable molecular switching of CaMKII action, referred to as autonomous activity (Chang et al., 2017; Lee et al., 2022; Tao et al., 2021), has made CaMKII an attractive candidate for a “memory molecule” in neuronal signaling (Lisman et al., 2002; Crick, 1984; Rossetti et al., 2017).
Other proteins, including kinases that are activated downstream of Ca2+ signaling, like PKC (Bear et al., 2018; Colgan et al., 2018), are proposed to be involved in synaptic plasticity, by altering cytoskeletal structure and receptor trafficking cycles. On the other hand, CaMKII is terrifically abundant in neurons and may have roles in addition to its enzymatic activity (like bundling actin; Okamoto et al., 2007; Kim et al., 2016). Particularly intriguing is the possibility that CaMKII alone could induce plasticity, in the absence of a substantial calcium signal, if it could be activated by a parallel means. To understand the specific role and biophysical mechanisms of CaMKII in calcium-triggered plasticity and memory, we sought to find a way to specifically activate CaMKII without calcium. To do this without the use of chemical activators or perturbing its structure by inserting large photo-responsive domains (Shibata et al., 2021), we exploited genetically-encoded unnatural amino acid mutagenesis, which we have previously used extensively in AMPA-type glutamate receptor ion channels in mammalian cells (Klippenstein et al., 2014; Poulsen et al., 2019) and more recently in CaMKII itself (Lučić et al., 2023). Using BzF (also known as Bpa), a well-studied UV-activated photocrosslinker (Nikić-Spiegel, 2020), we targeted the Calmodulin-binding footprint, aiming to irreversibly attach Calmodulin to CaMKII with light and maintain activity of the holoenzyme. Here we present in vitro characterization of this approach. Our results demonstrate that mutants that covalently trap Calmodulin on its cognate regulatory domains could offer spatial–temporal control of CaMKII activation and the activity of other targets of Calmodulin in mammalian cells.
2 RESULTS AND DISCUSSION
2.1 Characterization of CaMKIIL308BzF
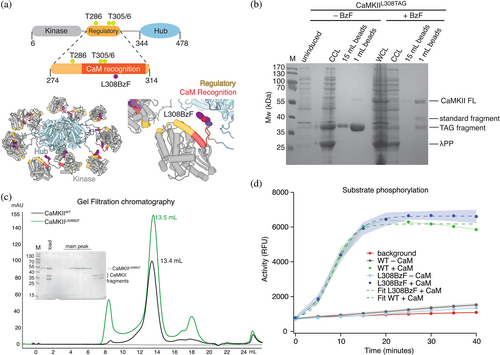
In order to activate CaMKII from here on referred to as CaMKII, by UV-induced crosslinking, we designed a mutant of CaMKII in which we replaced the leucine residue at position 308, in calmodulin-binding R3 element of the regulatory domain, with the photoreactive unnatural amino acid benzoyl-L-phenylalanine (BzF; Figure 1a). To achieve this, we first introduced an amber stop codon (TAG) in place of the leucine codon (CTG) by site-directed mutagenesis. To incorporate BzF in the CaMKII protein, we co-expressed CaMKII TAG-containing plasmid, together with an orthogonal tRNA, mutated to recognize the TAG codon, and aminoacyl tRNA synthetase (aaRS) which attaches BzF to the cognate tRNA (for detailed procedure see Section 4.2). Only E. coli that was grown in BzF-containing medium expressed full-length CaMKII (Figure 1b) from this construct, demonstrating that there was no substantial leaky expression of full-length “wild-type” CaMKII, caused by TAG stop codon readthrough. A minor protein band corresponding to full-length CaMKII was visible also in expression conditions without BzF. The rescue of full-length CaMKIITAG with BzF was not complete, because there was still a detectable fraction of truncated CaMKII, but the full-length CaMKIIL308BzF protein could be easily separated from the smaller fragments by size-exclusion chromatography. CaMKIIL308BzF had an elution volume similar to CaMKIIWT (Ve = 13.5 mL; Figure 1c), which corresponds well to the dodecameric holoenzyme elution profile of CaMKIIWT. Furthermore, the kinase activity of CaMKIIL308BzF toward a peptide-substrate, measured using ADPQuest kinase assay, was no different to CaMKIIWT (Figure 1d). The half-maximum time for peptide-substrate phosphorylation was about 8 min, similar to previous kinase activity of CaMKII measured in this way (Lučić et al., 2023). The reaction was complete in about 20 min. Given the 500-fold excess of substrate, this roughly corresponds to kcat (per kinase domain) of about 0.5 s−1 (500/1200 s) in our conditions. However, this estimate of kcat was measured at only one ratio of enzyme to substrate concentration and is therefore subject to a high uncertainty.
Both CaMKIIWT and CaMKIIL308BzF proteins were subjected to Intact Mass Spectrometry (Figure S1). Measured mass difference between the two proteins closely resembles the mass difference between residues L and BzF (∆m = 138 Da). Some impurities, of lower molecular weights, are detected in the mass spectra of CaMKIIL308BzF, possibly due to poor separation of the full length CaMKIIL308BzF from truncated fragments during size exclusion chromatography, which is not readily visible on an SDS-PAGE gel (Figure 1c). To our knowledge, CaMKII is the largest protein assembly successfully expressed with this technology.
We have also tested the rescue of CaMKIIL308TAG in mammalian HEK cells (Figure S2). In a cellular environment, both the rescued CaMKII with L308BzF and the truncated form (L308Stop) are produced. The rescue protocol requires further optimization in order to obtain more full-length CaMKII and less L308Stop protein product. For example, adding more BzF to the expression medium had a negative effect—it reduced the efficiency of the TAG codon rescue (Figure S2). However, the truncation at L308 should produce an enzyme that lacks Ca2+-stimulated activity, because the Calmodulin-binding site is disrupted by the truncation. Indeed, we can observe a lack of phosphorylation at residue T286 on L308Stop in HEK cells (Figure S2), whereas the phosphorylation on the full-length CaMKIIL308BzF still occurs.
2.2 UV-induced crosslinking of Calmodulin to CaMKIIL308BzF is Ca2+ dependent
Next, we examined whether CaMKIIL308BzF could be crosslinked to Calmodulin by UV light (Figure 2). The crosslinking induced by UV light is a covalent modification (Klán et al., 2013), unaffected by standard denaturing SDS-PAGE running conditions. Therefore, the formation of crosslinked complexes can be easily monitored with Coomassie staining of a polyacrylamide gel. To limit heat-induced denaturation and associated damage coming from the UV lamp exposure, we placed CaMKII samples in a passively cooled metal plate in a cold room (4°C) and treated the samples with light of 365 nm wave-length (see Section 4.6 for details). In order to mimic the high cellular concentration of CaMKII (Otmakhov & Lisman, 2012), we used 8 μM CaMKII and different concentrations of Calmodulin. Figure 2a shows a schematic representation of experiments performed in Figure 2b. Treatment of CaMKIIL308BzF with UV light in the presence of Calmodulin caused a nearly complete protein band shift on a denaturing SDS-polyacrylamide gel to higher molecular weight (Mw; from 55 to 70 kDa), corresponding to the combined mass of CaMKII monomer (55 kDa) and Calmodulin (17 kDa) (Figure 2b, lane 10). The depletion of vast majority of the 55 kDa band suggests stochiometric crosslinking, indicative of a tight interaction. The remaining signal detected in the monomeric band (around 5%, Figure 2b, lane 10) might lack the BzF residue at position L308, due to minor leaky expression of this construct, as seen in Figure 1b. The shift was absent when we used CaMKIIWT protein (Figure 2b, lane 4), showing that the cross-linking observed on the gel was specific to BzF residue at position 308 and Calmodulin. Furthermore, the crosslinking was absent if Calmodulin was previously stripped of Ca2+ by EGTA treatment (Figure 2b, lane 12), demonstrating that the complete reaction was dependent on the correct conformation of Calmodulin.
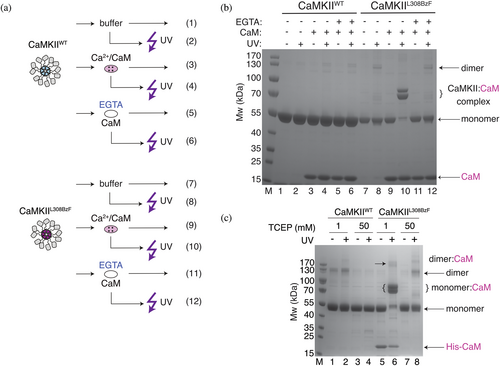
In CaMKIIWT samples, a band corresponding to the CaMKII dimer appeared after UV treatment. This band was absent when we diluted CaMKII in a buffer containing 50 mM TCEP, instead of our standard buffer which contained 1 mM TCEP (Figure 2c), indicative of the exposure of free cysteines, perhaps by sample heating or denaturation, which then allows nonspecific dimers of CaMKII to form. Although the double band appearing when CaMKIIL308BzF is crosslinked to Calmodulin could be explained by additional linking of the Cysteines in proximity of the residue 308 (Cys at position 280 or 289; Torres-Ocampo et al., 2020), which might be promoted more in CaMKIIL308BzF than in CaMKIIWT dimer, this double band persisted in 50 mM TCEP. Notably, following UV treatment of CaMKIIL308BzF alone (without Calmodulin), and in the presence of 50 mM TCEP (Figure 2c, lane 8), dimeric and higher order bands of CaMKIIL308BzF appeared, although this band is reduced in CaMKIIWT under the same conditions (Figure 2c, lane 4). If we interpret this observation as inter-subunit covalent crosslinking by the L308BzF residue, the question arises, how is this possible if the regulatory segment is always docked in basal conditions? These higher-order bands suggest that in the complete absence of CaM, the regulatory segment of CaMKII is mobile enough to contact other monomers, and is not persistently docked, which agrees well with previously published work on CaMKII dynamics (Hoffman et al., 2011). In this interpretation, in the presence of Calcium-Calmodulin, regulatory segments, although undocked, are cloaked by bound Calmodulin and cannot make intersubunit crosslinks. Even though higher-order bands were generally a minor fraction, we used 50 mM TCEP in most subsequent experiments to limit the formation of disulfide-bonded higher-order oligomers upon denaturation.
To further investigate the origin of the double band in CaMKII samples, we performed mass spectrometry analysis on CaMKIIL308BzF under several conditions (Figure S3). The analysis showed that, when exposed to UV light, CaMKII undergoes disulfide bridging on Cysteine 280 in the regulatory domain (Figure S3C). This was determined by treating CaMKII samples with an alkylation reagent (chloroacetamide; CAA) which binds only to free Cysteines resulting in carbamidomethyl modification (Giron et al., 2011; Goodman et al., 2018). We calculated the ratio of intensities between peptides with carbamidomethyl modification (free Cysteines) over unmodified peptides (disulfide bridged). We took 24 gel samples corresponding to four replicates of six gel bands (these six were drawn from four conditions; each condition is a lane run on the gel; Figure S3). Four of these samples (CaMKIIL308BzF, CaMKIIL308BzF UV treated, CaMKIIL308BzF incubated with Ca2+:Calmodulin, and CaMKIIL308BzF incubated with Ca2+:Calmodulin and UV treated—lower band from the doublet at 70 kDa) showed modification on Cys280 corresponding to Cys280 being free (Figure S3C), that is, not disulfide-bonded. However, we found that a portion of the sample of CaMKIIL308BzF incubated with Calmodulin and then treated with UV had a ratio of modified to base intensity (Imodified/Ibase) very close to 0 (due to very little or no modification; Figure S3B). The upper of the doublet band around 70 kDa in CaMKIIL308BzF incubated with Calmodulin and then UV treated, and the uncrosslinked band from the same condition (corresponding to CaMKII monomer), both show Imodified/Ibase equal to or close to 0, indicative of absence of carbamidomethyl modification. This absence of modification is likely caused by Cysteine 280 forming a disulfide bridge with another Cysteine in the vicinity. A similar pattern was found also for Cysteine 289, but the coverage for this peptide was not substantial, and it was not identified in all 4 replicates of each sample. We could also not confirm BzF incorporation in individual bands because of poor coverage. We did not test the double bands appearing at around 120 kDa corresponding to CaMKII dimers, but we suspect that they are also the result of similar disulfide bridging.
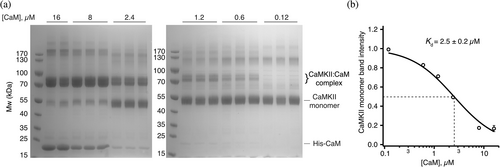
Using UV-induced cross-linking we were able to determine the binding constant of Calmodulin to CaMKIIL308BzF. A total of 8 μM CaMKIIL308BzF was incubated with different concentrations of Calmodulin (16, 8, 2.4, 1.2, 0.6, and 0.12 μM) in triplicate, and each reaction was exposed to UV, followed by SDS-PAGE analysis. By measuring CaMKII monomer band densitometry from Coomassie-stained gel, we found that UV-induced crosslinking of Calmodulin to CaMKII depletes the CaMKII monomer band, as a result of shifting to the Ca2+:Calmodulin:CaMKIIL308BzF complex, with KD of 2.5 ± 0.2 μM (Figure 3). Despite being measured in a completely different way, and despite the very strong dependence of association on calcium concentration, this affinity is not starkly different from the previously reported affinity (0.6 μM) of Calmodulin to CaMKIIα from ITC measurements (Rellos et al., 2010), suggesting that the incorporation of BzF at position 308 does not alter the association of the CaMKII:CaM complex much.
2.3 Calmodulin binding does not change oligomeric state of CaMKIIL308BzF
It has been proposed that binding of Ca2+:Calmodulin during CaMKII activation causes the release of subunits from the dodecameric CaMKII holoenzyme (Bhattacharyya et al., 2016; Karandur et al., 2020; Stratton et al., 2014). We wanted to directly test the effect of Ca2+:Calmodulin binding alone to CaMKII holoenzymes on the oligomeric state of CaMKIIL308BzF. The advantage of our approach is that we get almost 100% of CaMKIIL308BzF crosslinked to Calmodulin upon UV exposure, so any effect that Calmodulin might exert on oligomeric state of CaMKII should not be masked by incomplete binding. To this end, we employed mass photometry (Figure 4), a single molecule technique that measures light scattered from mobile particles in solution as they make transient contact a coverslip, enabling calculation of their Mw (Young et al., 2018). Detected particles are described as counts on the y-axis of the graphs in Figures 4, S4, and S5. The measurements were performed in triplicate (Figures S4 and S5), demonstrating good reproducibility between measurements. We tested three different concentrations of CaMKII (400, 100, and 10 nM). CaMKII formed dodecameric holoenzymes even at the lowest concentration, 10 nM (Figure S4D). In all measured conditions and at all concentrations, a small peak corresponding to monomer-dimer CaMKII was detected, assuring that the instrument can indeed detect smaller particles. In some of the measurements, a peak at negative Mw values appeared, which corresponds to particle unbinding events. However, this peak was, if there, always a minor fraction (2%–5%). We also reported sigma values for each peak, which correspond to peak width and inform us about the distribution of the particles' sizes.
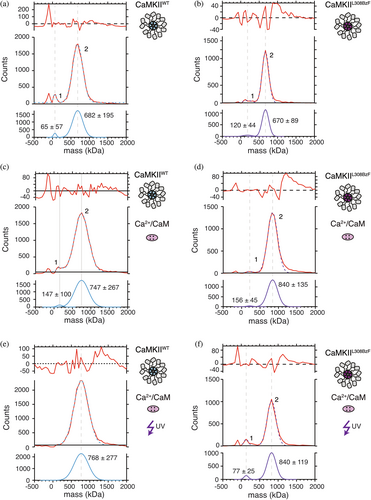
After mass photometry, each sample was run on SDS-PAGE to validate the reactions (Figure S4F). Two different batches of purified CaMKIIL308BzF were compared, as well as two different Calmodulins (self-made and commercial, purchased from Calbiochem). The electrophoresis results were similar between all combinations of CaMKII and Calmodulin (Figure S4F).
We first measured isolated CaMKIIWT protein (Figure 4a), and compared it to either CaMKIIWT incubated with Ca2+:Calmodulin in the absence (Figure 4c) or presence of UV light (Figure 4e), as well as to CaMKIIL308BzF alone (Figure 4b), CaMKIIL308BzF incubated with Ca2+:Calmodulin in the absence (Figure 4d) or presence of UV light (Figure 4f). The concentration of all CaMKII proteins was initially set to 8 μM and Calmodulin to 8 μM. Selected samples were then treated with UV light, and the concentration of all samples was diluted to 400 nM (CaMKII monomer concentration) in the measured drop on the instrument, because particles were too dense at higher concentrations, so the instrument could not distinguish them from one another.
Consistent with previous work (Lučić et al., 2023; Torres-Ocampo et al., 2020), particles of isolated CaMKIIWT (Figure 4a) gave a major peak with estimated Mw corresponding to dodecameric holoenzymes (Mw = 682 kDa), together with a smaller peak (around 1%–2%) corresponding to monomer-dimer Mw. Incubation of CaMKIIWT with Ca2+:Calmodulin (Figure 4c) resulted in the appearance of a major peak with Mw = 747 kDa and a smaller shoulder peak (around 1%–2%) at the lower Mw, similar to that one seen in Figure 4a. The major peak corresponds to partially Calmodulin-bound dodecamers (12 × 55 kDa + 5 × 16.4 kDa = 742 kDa). We cannot exclude that other combinations of CaMKII:CaM would give a similar Mw (e.g., a decamer with 10 CaM bound would be ~720 kDa). Nonetheless, there is no major monomer-dimer peak detected upon incubation with Calmodulin. Treatment of Ca2+:Calmodulin and CaMKIIWT mixture with UV resulted in a single peak of Mw = 768 kDa, which would correspond to one holoenzyme of CaMKII with 6 Ca2+:Calmodulin bound (12 × 55 kDa + 6 × 16.4 kDa = 758 kDa) (Figure 4e), similar to the untreated CaMKIIWT sample.
Particles coming from CaMKIIL308BzF alone were measured to have similar Mw (670 kDa) like the CaMKIIWT (Figure 4b) as expected. Again, a small peak (1%–2%) corresponding to CaMKII dimer size (120 kDa) was detected. Detected size of CaMKIIL308BzF in the presence of Calmodulin, but without UV treatment (Figure 4d; Mw = 840 kDa), corresponds to fully Calmodulin-bound CaMKIIL308BzF (12 × 55 kDa + 12 × 16.4 kDa = 855 kDa). This discrepancy between particle sizes of CaMKIIWT and CaMKIIL308BzF incubated with Calmodulin could come from different Calmodulin concentrations used in these experiments (2 μM Calmodulin with CaMKIIWT, and 8 μM Calmodulin with CaMKIIL308BzF). However, when we compared CaMKIIWT (Figure 4c, e) to CaMKIIL308BzF using the same Calmodulin (Figure S5E, F), the difference was absent. A small dimeric peak (Mw = 156 kDa) preceding the main peak (Figure 4d) is still in small abundance (around 1%–2%) similar to the one measured in the condition in Figure 4b, and could have Calmodulin bound to it (55 kDa × 2 + 16.4 kDa × 2 = 143 kDa). It is expected that dimers which dissociate from holoenzymes due to concentration-dependent dissociation are still capable of binding to Calmodulin. Irreversible crosslinking of CaMKIIL308BzF:Calmodulin mixtures with UV light did not further change the stoichiometry of CaMKIIL308BzF holoenzymes (Figure 4f; Mw = 840 kDa).
Perhaps contrary to expectation, under the conditions used here, we failed to detect particles corresponding in size to dissociated CaMKII subunits dependent on Calmodulin presence. It is possible that holoenzymes did not dissociate because we omitted ATP from these measurements. Therefore, trans-autophosphorylation of the regulatory domain, which might be necessary for the disassembly of holoenzymes (Bhattacharyya et al., 2016; Karandur et al., 2020), was not promoted. However, these results show that Calmodulin binding to CaMKII in and of itself, does not break up the CaMKII holoenzyme, even when the binding is stochiometric, and irreversible like in the case of UV-treated CaMKIIL308BzF:Ca2+:Calmodulin mixture (Figure 4f).
2.4 Ca2+ is dispensable for the activity of constitutively calmodulin bound CaMKII
Binding of Ca2+-bound Calmodulin to CaMKII is the first step in the activation of CaMKII, and it is necessary to relieve the inhibition posed by the regulatory domain, which otherwise obstructs the substrate binding cleft in naïve CaMKII (Rellos et al., 2010; Shifman et al., 2006). In order to bind to CaMKII, we would expect that Calmodulin should adopt the correct conformation, which is obtained by Ca2+ binding (Shifman et al., 2006). Only when Ca2+:Calmodulin is bound to the regulatory domain, can CaMKII trans-autophosphorylate itself at position T286, at which point CaMKII no longer needs Ca2+:Calmodulin because phosphorylation at T286 prevents rebinding of the regulatory domain to the substrate binding cleft, and CaMKII can now freely engage its substrates (Rellos et al., 2010). This Ca2+:Calmodulin-independent state is known as autonomous activity of CaMKII (Lee et al., 2022).
The mutant CaMKIIL308BzF allowed us to ask whether CaMKII can be switched into autonomous activity (synonymous with ongoing T286 phosphorylation) in very low calcium. To do this, we performed experiments in high, intermediate, and very low free calcium concentrations (2 mM, 26 nM, and 540 pM; using 7 and 40 mM EGTA for the second and third conditions, free Calcium was calculated with MaxChelator). These results are shown in Figures 5 and S3. For clarity, Figure 5a shows schematic representations of the experiments performed in Figure 5b (likewise in Figure S6). Incubation of mixtures containing Ca2+:Calmodulin and CaMKIIL308BzF with EGTA, prior to UV treatment, was sufficient to abolish CaMKIIL308BzF:Calmodulin crosslinking (Figures 2b, 5b; top panel, lane 10; Figure S6B, top panel, lane 14). Minor phosphorylation signals could still be detected, albeit at about 20–30× less intensity, on these CaMKIIL308BzF species, as well as on CaMKIIWT without UV treatment (Figure 5b, middle panel, lane 10, lanes 3 and 4; Figure S6B lanes 3 and 4) but this was not consistent (Figure S6B, middle panel, lanes 14 and 15). These results indicate that there is almost no interaction between Calmodulin and regulatory domain of CaMKII in the presence of Ca2+ chelating agent, and therefore, at most a very small portion of trans-autophosphorylation. However, if we first treated the mixtures of Ca2+:Calmodulin and CaMKIIL308BzF with UV, and then added EGTA (Figure 5b, lane 12; Figure S6B, lane 16), the phosphorylation signal was strong and indistinguishable to samples that were never exposed to EGTA (Figure 5b, lane 9; Figure S6B lane 12). This observation means that the crosslinking of Ca2+:Calmodulin to CaMKIIL308BzF either prevents stripping of Ca2+ from Calmodulin or simply enables tight binding of Calmodulin to the regulatory domain of CaMKII, perhaps independent of Calmodulin conformation. In any case, this crosslinking switched CaMKII to calcium-independent phosphorylation of T286, resembling autonomous activity.
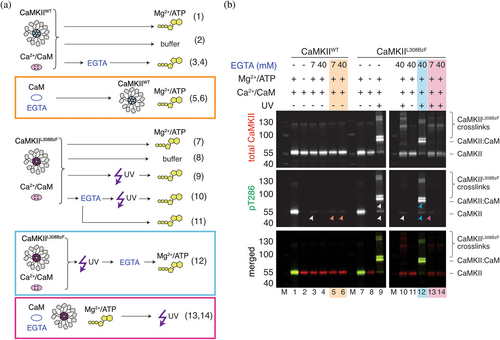
Additionally, pre-incubation of Calmodulin with CaMKII might enhance Calmodulin affinity for Ca2+, as has been previously reported (Shifman et al., 2006), making it more difficult to chelate Ca2+ ions with EGTA. Incubation of premixed CaMKII and Ca2+:Calmodulin with a high amount of EGTA ([EGTA]:[Ca2+] = 20) prior to UV treatment, still allowed for residual phosphorylation on T286 in mixtures that were subsequently UV treated (Figure 5B, middle panel, lane 10), but if the mixture was not subsequently subjected to UV, the phosphorylation signal was completely abolished (Figure 5b, middle panel, lane 11). This result further indicates that our UV-sensitive mutant can capture a tiny residual proportion of tightly-bound CaM in nominally ultra-low calcium and that this is sufficient to allow phosphorylation (note, pre-stripping CaM of calcium was more effective at stopping phosphorylation). Incubation of Ca2+:Calmodulin with EGTA prior to addition to CaMKIIL308BzF was sufficient to prevent Calmodulin:CaMKIIL308BzF crosslinking (Figure 5B, top panel, lanes 13 and 14; Figure S6B, top panel, lanes 17 and 18), although it allowed UV-dependent formation of higher-order CaMKII oligomers, as in Figure 2, likely due to interaction of regulatory domain of CaMKII of one monomer to the regulatory or kinase domain of CaMKII of adjacent monomers (Chao et al., 2010; Rocco-machado et al., 2022). This result further indicates that domains within the dodecamer are mobile, making dynamic inter-subunit contacts, and also perhaps that the regulatory segment is not permanently docked on the substrate binding site of its parent subunit (Hoffman et al., 2011).
The phospho-signal was absent when Ca2+:Calmodulin was pre-incubated with high EGTA (20-fold excess, Figure 5b, middle panel, lane 14; Figure S6B, middle panel lane 18), due to efficient chelation of Ca2+. On the other hand, pre-incubation of Ca2+:Calmodulin with EGTA in 3.5-fold excess of Ca2+ was not sufficient to completely abolish phosphorylation of CaMKIIL308BzF (Figure 5b, middle panel, lane 13; Figure S6B, middle panel, lane 17). Similar results were obtained with the wt protein (Figure 5b, middle panel, lanes 3 and 4; Figure S6B, middle panel, lanes 3 and 4), although in this case incubation of CaMKIIWT with the same Ca2+:Calmodulin:EGTA mixture (see Section 4.9 for details) with a 20-fold excess of EGTA to Ca2+ still did not completely abolished the phosphorylation signal (Figure 5b, middle panel, lanes 5 and 6; Figure S6B, middle panel, lane 5), although it was reduced 80-fold, compared to EGTA untreated sample (Figure 5b, middle panel, lane 1). However, we cannot exclude the possibility that residual binding of free Mg2+ ions (we had 15 and 1.3 mM free Mg2+ in the presence of 7 and 40 mM EGTA, respectively; calculated with MaxChelator) to Calmodulin in these reactions could somehow mediate its proper folding and therefore binding to CaMKII. The small discrepancy between CaMKIIWT and CaMKIIL308BzF levels of phosphorylation in these reactions might arise from a modest difference in Calmodulin affinity between wt and L308BzF mutant, in which wt residue L on position 308 makes residual binding of Calmodulin possible and sufficient to drive low phosphorylation of T286 (80× lower than in EGTA untreated sample). Since the phosphorylation signal was completely absent in reactions where either ATP:Mg2+ was omitted from the reaction (Figure 5b lanes 2 and 8, Figure S6B lanes 2 and 11) or Ca2+:CaM (Figure S6B lanes 7 and 8), it is clear that residual phosphorylation occurring at ultra-low calcium is not a consequence of pre-existing phosphorylation on CaMKII which might have occurred in E. coli during expression.
3 CONCLUSION
Using a photoactive unnatural amino acid as a tool for probing CaMKII:Calmodulin interaction, we could show that Calmodulin binding to CaMKII alone does not cause disassembly of the CaMKII holoenzyme (Figure 4). The CaMKIIL308BzF mutant, which maintained the stoichiometric binding characteristic of wild-type CaMKII, allowed the irreversible attachment of Calmodulin with UV-induced crosslinking. This crosslinked complex showed similar activity to the wild-type kinase, even under conditions of negligible free calcium. Our activity assays also revealed that a residual auto-phosphorylation of CaMKII is still present even when Calmodulin:CaMKIIL308BzF crosslinking is undetectable on western blot, indicating that a small amount of Calmodulin might remain constitutively bound to CaMKII. Finally, cross-linking of higher-order inter-subunit CaMKII complexes by L308BzF in the absence of CaM demonstrated that the regulatory segment is mobile in basal conditions.
Following from these observations, the CaMKIIL308BzF mutant could be a promising candidate for irreversible photo-activation of CaMKII in cells. However, the CaMKII L308Stop truncation may behave differently from native CaMKII in other ways, particularly as it lacks the hub domain. Nonetheless, this mutant might find utility as a complement to previous functional mutants of CaMKII that promoted constitutive activity, which also lacked the hub domain, and alternative methods for controlling CaMKII with light (Parra-bueno et al., 2017; Shibata et al., 2021). Such a tool might be used for acute activation of CaMKII, in order to promote processes like long-term potentiation of synaptic transmission in neurons with a high temporal precision. This application is particularly interesting in light of the recent data suggesting that CaMKII modification in the regulatory domain triggers plasticity, even in the absence of enzymatic function (Chen et al., 2023; Tullis et al., 2023).
4 MATERIALS AND METHODS
4.1 Expression constructs
Human wild-type CaMKII (CaMKIIWT), with a 30 residue linker between regulatory and hub domains, was gene optimized for expression in E. coli, and the first 5 residues of the CaMKII protein were deleted (Chao et al., 2011). A hexa-his tag was placed on the N-terminus, followed by TEV cleavage site, and synthesized by GenScript in pET28b + vector, flanked by NcoI and HindIII restriction sites. CaMKIIL308BzF was cloned over CaMKIIWT background using TAG-containing primers, by site-directed-mutagenesis. Human CaMKIIWT-mScarlet used for expression in mammalian cells was cloned by fusing the human CaMKIIWT gene purchased from Addgene (plasmid# 23408, ref; Johannessen et al., 2011) to mScarlet purchased from Addgene (plasmid# 85054, ref; Bindels et al., 2016), and the L308TAG site was introduced by site-directed-mutagenesis. Human Calmodulin with an N-terminal His8 tag, followed by a TEV cleavage site, was codon optimized for expression in E. coli in pET28b + plasmid, and gene synthesized by GenScript. Lambda protein phosphatase (LPP) was purchased from Addgene (plasmid #79748; Albanese et al., 2018). The plasmid containing TEV protease was a kind gift from Prof. Dr. Thomas Leonard (Max Perutz Laboratories, Vienna, Austria). The plasmid containing aminoacyl synthetase (aaRS) and tRNA from M. jannaschii (Young et al., 2010) in pEVOL backbone, was a kind gift from Prof. Dr. Thomas Söllner (BZH, Heidelberg, Germany). Human Calmodulin with an N-terminal His8 tag, followed by a TEV cleavage site, was codon-optimized for expression in E. coli and gene synthetized (GenScript) using pET28b + vector as a backbone. The BzF tRNA and RS for mammalian cell expression were a kind gift from Thomas Sakmar (Rockefeller; Ye et al., 2008).
4.2 CaMKII expression
Expression of CaMKIIWT was done in BL21 E. coli cells, grown in LB medium. To aid solubility, CaMKIIWT (Kanamycin resistance) was co-expressed with LPP (Spectinomycin resistance). Cells were grown in LB medium at 37°C/200 rpm until OD600 reached 1.2–1.5. Expression of both genes was induced by adding 0.4 mM IPTG, and the medium was supplemented with 0.5 mM MnCl2 (co-factor of LPP). Protein expression was continued overnight at 20°C.
Expression of CaMKIIL308BzF was performed by co-expression of CaMKIIL308TAG (Kanamycin resistance), LPP (Spectinomycin resistance), and a plasmid containing orthogonal aaRS and tRNA from M. jannaschii (Chloramphenicol resistance; Young et al., 2010) in E. coli BL21 cells. Cells were grown at 37°C/200 rpm in LB medium until they reached OD600 = 0.6. The medium was then supplemented with 1 mM BzF (BACHEM, product #4017646), and cells continued to grow for another 30 min. The temperature was then lowered to 30°C, and the aaRS and tRNA expression was induced by adding 15 mM arabinose. The cells continued to grow until the OD600 reached 1.2–1.5, the temperature was lowered to 20°C, and CaMKIIL308TAG and LPP expression was induced by addition of 0.4 mM IPTG. The medium was supplemented with 0.5 mM MnCl2 and the cultures were grown overnight at 20°C.
4.3 Calmodulin expression
Human Calmodulin (Kanamycin resistance) expression was performed in Rosetta DE3 cells (Chloramphenicol resistance) in a TB medium, supplemented with 5 mM CaCl2. Protein expression was induced at OD600 = 1.8 by 1 mM IPTG. Cells were harvested after growing for 5 h at 37°C/200 rpm.
4.4 CaMKII purification
Cell pellets expressing either CaMKIIWT or CaMKIIL308BzF were lysed in lysis buffer (50 mM Tris pH 8, 300 mM NaCl, 20 mM imidazole, 1 mM TCEP) supplemented with 0.02 mg/mL DNaseI, 5 mM MgCl2, 0.5 mg/mL lysozyme and 1 mM PMSF. The lysates were additionally passed through a cell disruptor two times before the lysates were cleared of cell debris by centrifugation (16k rpm, 4°C). Both constructs have an N-terminal His6 tag, which was used for affinity purification with 5 mL NiNTA column (GE Healthcare), previously equilibrated in NiA buffer (50 mM Tris pH 8, 300 mM NaCl, 20 mM imidazole, 1 mM TCEP). The column was then extensively washed (100 mL of NiA buffer, 50 mL of NiA buffer with 50 mM imidazole, 50 mL of NiA buffer with 80 mM imidazole), and eluted in a gradient elution from 80 mM imidazole to 1 M imidazole (50 mM Tris pH 8, 300 mM NaCl, 1 mM TCEP, 1 M imidazole) over 10 column volumes. Peak fractions were pooled, imidazole concentration lowered to less than 100 mM with NiA, and the sample was incubated with 0.01 mg/mL final TEV (made in-house) in order to cleave off the His6 tag. On the following day, the cleaved protein was concentrated to 500 μL, using 50 kDa cut-off concentrator (4 mL Amicon R Ultra), and injected into Superose 6 10/300 column (GE Healthcare) previously equilibrated in size exclusion (SEC) buffer (25 mM Tris pH 8, 250 mM NaCl, 1% glycerol, 1 mM TCEP). Elution profile of CaMKIIL308BzF comprised of three peaks: the first one which corresponded to Mw of dodecameric CaMKII (around 650 kDa), the second one which corresponded to ~40 kDa CaMKII fragment, which is usually present during CaMKII purification from E. coli, and the third one corresponding to ~35 kDa L308Stop fragment. The peak corresponding to dodecameric CaMKII was pooled and further concentrated to approximately 2 mg/mL. We obtained around 1–2 mg of protein per 2 L of E. coli culture.
The small-scale test purification reported in Figure 1b was performed using a modified version of the protocol described above. Upon clearance of cell lysate from 0.25 L of cell pellets, the lysate was then incubated with 1 mL of Co2+ beads. After 2 h incubation with the beads (rolling in a falcon tube at 4°C), the beads were spun down, the supernatant removed and beads resuspended in 15 mL of wash buffer (NiA buffer). After a few cycles of bead washing, the protein was cleaved from the beads, using TEV protease, overnight, rolling at 4°C. Cleaved protein was run on an SEC column, as described above.
4.5 Calmodulin purification
Cell pellets from 4 L of bacterial cultures were lysed in 100 mL of lysis buffer (50 mM Tris pH 8, 150 mM NaCl, 1 mM PMSF, 0.02 mg/mL DNaseI, 5 mM MgCl2, and 0.5 mg/mL lysozyme) for 1 h stirring at 4°C, followed by sonication on ice (6 × 25 s, 5 cycle, 50% power, 1 min pause between cycles). Cell lysates were cleared by centrifugation (16k rpm, 4°C for 30 min), and the supernatant was loaded on a 5 mL NiNTA column (GE Healthcare), previously equilibrated in NiA buffer (50 mM Tris pH 8, 150 mM NaCl, 5 mM MgCl2 and 20 mM imidazole). His-tagged Calmodulin was eluted in a gradient elution, using NiB buffer (50 mM Tris pH 8, 150 mM NaCl, 5 mM MgCl2, and 1 M imidazole). Peak fractions were pooled and incubated with 150 μL of 2 mg/mL TEV (made in-house) overnight. To optimize the cleavage conditions, imidazole concentration was lowered to under 100 mM, and 1 mM TCEP was added to the cleavage reaction. The purification was continued on the following day by loading the overnight sample onto a 1 mL Q column (GE Healthcare). To enable binding to the column, first, the salt concentration was lowered to about 40 mM NaCl by adding QA buffer (20 mM HEPES pH 7, 25 mM NaCl, 5 mM CaCl2). Calmodulin was eluted from the column by gradient elution, using QB buffer (20 mM HEPES pH 7, 1 M NaCl, 5 mM CaCl2). Fractions were run on an SDS-PAGE gel to determine the peak fractions, because the 280 nm signal coming from Calmodulin is too low, as Calmodulin does not contain any Trp residues. The peak fractions were pooled and concentrated to 200 μL. Calmodulin concentration was determined using Bradford assay and found to be around 0.8 mg/mL (47 μM).
4.6 UV-induced crosslinking
To drive crosslinking of CaMKIIL308BzF to Calmodulin, we used a UV LED that emits UV light of 365 nm wavelength (Opsytec Dr. Gröbel). Samples were placed in a cold metal plate (made in-house) in a cold room, to avoid damage caused by excess heating from the UV light. Each sample was illuminated 5 times using 15 1 s pulses of UV light, with a 1 s break in between each pulse. This was repeated 5 times per sample with a 1-min break between each series of pulses. Samples were in SEC buffer, supplemented with either 1 mM TCEP or 50 mM TCEP to avoid non-specific crosslinking due to oxidation of surface exposed Cysteines. The samples were then run on a 4%–12% gradient Bis-Tris gel (ThermoScientific), and stained with Coomassie dye (FastGene Q Stain from Nippongenetics).
4.7 Determination of Calmodulin binding constant
4.8 Mass photometry
Mass photometry was performed on Refeyn OneMP mass photometer. Coverslips (Marienfeld high precision glass coverslips, 24 × 50 mm #1.5H, 170 ± 5 μm; product # 0107222) were cleaned following the manufacturer's instructions. The coverslips were rinsed sequentially with purified miliQ H2O, 100% EtOH, 100% isopropanol, miliQ H2O, 100% EtOH, and miliQ H2O several times, then dried under a clean stream of nitrogen.
The samples of CaMKIIWT were prepared by diluting 8 μM CaMKIIWT with or without 2 μM self-made Calmodulin, with or without UV exposure in buffer containing 25 mM Tris pH 8, 250 mM NaCl, 1% glycerol, 1 mM TCEP to obtain 1.6 μM CaMKIIWT. The samples of CaMKIIL308BzF were prepared by diluting 8 μM CaMKIIL308BzF with or without 8 μM His-Calmodulin (Calbiochem), with or without UV exposure in buffer containing 25 mM Tris pH 8, 250 mM NaCl, 1% glycerol, 1 mM TCEP to obtain 1.6 μM CaMKIIWT. Samples were initially incubated for 5 minutes at room temperature, before the first image acquisition. Each sequential measurement meant that the sample spent additional 1–2 min incubating before it could be recorded again, for each triplicate. Images were collected in an area of 3 × 10 μm, except for wild-type CaMKII in Figure 4a, c, e which were done with the large view (10 × 10 μm). CaMKIIL308BzF 8 μM was also diluted to 0.4 μM and 0.04 μM. The recordings were made by diluting 5 μL of stock samples (1.6, 0.4, or 0.04 μM) in 15 μL of buffer, directly on the cover slide. The final concentration of CaMKII (monomer concentration) in the drop was either 400 nM (for both CaMKIIWT or CaMKIIL308BzF), 100 nM or (CaMKIIL308BzF) 10 nM (CaMKIIL308BzF). The counts obtained for each measurement were fitted to a Gaussian fit using IgorPro9 software Multipeak fitting package.
4.9 Kinase activity assay
The following master mixes were set up:
Master mix 1 (MM1) containing 4.4 μM CaMKIIWT, 2.2 μM self-made Calmodulin, 2.2 mM CaCl2 in SEC buffer (with freshly added 50 mM TCEP), master mix 2 (MM2) containing 4 μM Calmodulin, 4 mM CaCl2, 14 mM EGTA (incubated for 5 min at 25°C to allow chelation of Ca2+), master mix 3 (MM3) containing 4 μM Calmodulin, 4 mM CaCl2, 80 mM EGTA (incubated for 5 min at 25°C to allow chelation of Ca2+), master mix 4 (MM4) containing 4.4 μM CaMKIIL308BzF, 2.2 μM self-made Calmodulin, 2.2 mM CaCl2 in SEC buffer (with freshly added 50 mM TCEP). Activation with ATP:Mg2+ was done at 37°C for 10 min, in all reactions where ATP:Mg2+ was added.
Reactions containing CaMKIIWT were set up, to obtain approximately final [CaMKII] = 4 μM, final [CaM] = 2 μM and [CaCl2] = 2 mM, as follows:
Lane 1–9 μL of MM1 activated by adding 1 μL of 1 mM ATP/ 200 mM MgCl2 to get final 100 μM ATP and 20 mM MgCl2, lane 2–9 μL of MM1 with 1 μL of SEC buffer, lane 3–8 μL of MM1 incubated with 1 μL of 70 mM EGTA (final [EGTA] = 7 mM) for 5 min at 25°C, followed by activation by adding 1 μL of 1 mM ATP/ 200 mM MgCl2 to get final 100 μM ATP: 20 mM MgCl2, lane 4–8 μL of MM1 incubated with 1 μL of 400 mM EGTA for 5 min at 25°C (final [EGTA] = 40 mM, note, final CaMKII 3.5 μM in this reaction), followed by activation by adding 1 μL of 1 mM ATP/ 200 mM MgCl2 to get final 100 μM ATP: 20 mM MgCl2, lane 5–5 μL of MM2 with 4 μL of CaMKIIWT (9.9 μM) for 5 min at 25°C, then activated by adding 1 μL of 1 mM ATP/ 200 mM MgCl2 to get final 100 μM ATP: 20 mM MgCl2, lane 6–5 μL of MM3 with 4 μL of CaMKIIWT (9.9 μM) for 5 min at 25°C then activated by adding 1 μL of 1 mM ATP/ 200 mM MgCl2 to get final 100 μM ATP: 20 mM MgCl2.
Reactions containing CaMKIIL308BzF were set up, to obtain final [CaMKII] = 4 μM, final [CaM] = 2 μM and [CaCl2] = 2 mM, as follows:
Lane 7–9 μL of MM4 activated by adding 1 μL of 1 mM ATP/ 200 mM MgCl2 to get final 100 μM ATP and 20 mM MgCl2, lane 8–9 μL of MM1 with 1 μL of SEC buffer, lane 9–9 μL of MM4 treated with UV and then activated by adding 1 μL of 1 mM ATP/ 200 mM MgCl2 to get final 100 μM ATP and 20 mM MgCl2, lane 10–8 μL of MM4 with 1 μL of 400 mM EGTA for 5 min at 25°C, followed by UV treatment and then addition of 1 μL of 1 mM ATP/ 200 mM MgCl2 to get final 100 μM ATP and 20 mM MgCl2 (note, final CaMKII 3.5 μM in this reaction and lanes 11 and 12).
Lane 11–8 μL of MM4 with 1 μL of 400 mM EGTA for 5 min at 25°C, without UV treatment, followed by the addition of 1 μL of 1 mM ATP/ 200 mM MgCl2 to get final 100 μM ATP and 20 mM MgCl2, lane 12–8 μL of MM4 was first treated with UV to obtain CaMKII:CaM crosslinks, then incubated with 1 μL of 400 mM EGTA for 5 min at 25°C, followed by activation with 1 μL of of 1 mM ATP/ 200 mM MgCl2 to get final 100 μM ATP and 20 mM MgCl2, lane 13–5 μL of MM2 with 4 μL of CaMKIIL308BzF (9.9 μM) for 5 min at 25°C, then activated by adding 1 μL of 1 mM ATP/ 200 mM MgCl2 to get final 100 μM ATP: 20 mM MgCl2, and then treated with UV, lane 14–5 μL of MM3 with 4 μL of CaMKIIL308BzF (9.9 μM) for 5 min at 25°C, then activated by adding 1 μL of 1 mM ATP/ 200 mM MgCl2 to get final 100 μM ATP: 20 mM MgCl2, and then treated with UV.
The kinase assays were repeated with newly purified CaMKIIWT and CaMKIIL308BzF, using His-Calmodulin from Calbiochem. The master mixes were set up like before, but this time using equimolar amounts of CaMKII and His-Calmodulin. The following master mixes were set up: master mix 1 (MM1) containing 4.4 μM CaMKIIWT, 4.4 μM His-Calmodulin, 2.2 mM CaCl2 in SEC buffer (with freshly added 50 mM TCEP), master mix 2 (MM2) containing 8 μM His-Calmodulin, 4 mM CaCl2, 14 mM EGTA (incubated for 5 min at 25°C to allow chelation of Ca2+), master mix 3 (MM3) containing 8 μM Calmodulin, 4 mM CaCl2, 80 mM EGTA (incubated for 5 min at 25°C to allow chelation of Ca2+), mater mix 4 (MM4) containing 4.4 μM CaMKIIL308BzF, 4.4 μM self-made Calmodulin, 2.2 mM CaCl2 in SEC buffer (with freshly added 50 mM TCEP). The reactions were set up like those described above. Conditions in each lane are described in Figure S3 legend. Controls for basal CaMKII activity (no Calmodulin) were added (lanes 7 and 8).
4.10 Kinase activity assay with peptide-substrate
Peptide-substrate (Syntide from GenScript) phosphorylation was measured using ADPQuest according to the manufacturer instructions. Reactions were set up in a 96-well plate and the kinase reaction was monitored using a TECAN plate reader. Briefly, activity of 100 nM CaMKII toward 50 μM Syntide, incubated with 100 μM ATP/200 μM MgCl2 and 1.5 μM Ca2+:Calmodulin, was measured for 40 min, at 37°C. Recordings were made every 5 min.
4.11 Western blot detection on samples from in vitro kinase assays
First, 500 ng of CaMKII protein was loaded per lane of a pre-casted 4%–12% gradient BisTris gel (ThermoScientific) to allow protein separation, followed by transfer to pre-activated (in 100% MeOH) PVDF membrane (Millipore) for 2 h at 50 V at 8°C, using Criterion Blotter (BioRad) in Twobin's transfer buffer with 20% MeOH. Membranes were then blocked in Tris Buffer Saline (TBS) with 0.1% Tween and 5% Milk (Sigma Aldrich) for 1 h shaking at room temperature. The membranes were incubated with two primary antibodies at the same time (mouse anti-human pan CaMKII G69 from CellSignaling (product #50049S), and rabbit anti-pT286 from CellSiganilng (product #12716S)), in 5% milk/TBS-T over-night at 4°C. On the following day, the membranes were washed 3× in TBS-T and incubated with secondary antibodies (fluorescently labeled IRDye 800CW Goat-anti Rabbit IgG and IRDye 680LT Goat-anti Mouse IgG from LiCor) for 1 h shaking at room temperature, followed by 3× washing in TBS-T. This allowed simultaneous detection of both total CaMKII (excited with 700 nm wavelength) and phosphorylation at T286 (excited with 800 nm wavelength) signals, using a LiCor imager.
4.12 HEK cell transfection and lysis
HEK 293 cells (ACC-305) were purchased from Leibniz Institute DSMZ (German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig). In-house testing for mycoplasma yielded a negative result.
A total of 1 million cells were seeded per one well of a 6-well dish. The cells were transfected on the following day using polyethylenimine (PEI), in a DNA:PEI ratio of 1:3. Total of 3 μg of DNA with CaMKIIL308TAG:tRNA:RS ratio of 4:1:1 (2 μg of CaMKIIL308TAG, 0.5 μg of tRNA and 0.5 μg of RS) was mixed with 9 μL of PEI in 250 μL prewarmed optiMEM. After 4–5 h the media was changed to remove the transfection regent and to add fresh media supplemented with BzF (1, 5, or 10 mM). Cells were harvested after 24–48 h, by adding 1 mL of ice-cold DPBS per well, and centrifugation at 2000 rpm for 5 min in a cooled table-top centrifuge. The cell pellets were then lysed in 100 μL of ice cold lysis buffer (50 mM Tris pH 8, 150 mM NaCl, 1% Triton X 100, 1 mM TCEP, 1 μM Pepstatin A, 10 μM Leupeptin, 0.3 μM Aprotinin, 1 mM PMSF, 5 mM NaF, 0.02 mg/mL DNaseI and 5 mM MgSO4) on ice, for 20 min, with vortexing every 5 min. The lystaes were then cleared by high-speed centrifugation for 15 min in a cooled tabletop centrifuge. Supernatants, containing CaMKII were subjected to Western Blot analysis.
4.13 Western blot detection on HEK cell lysates
A total of 21 μL of each lysate was combined with 7 μL of 4 × SDS loading dye and incubated at 95°C for 2 minutes to aid protein denaturation. A total of 20 μL of each sample was loaded on 4%–12% pre-casted gel (Thermo Scientific), and ran for 1 h 20 min at 180 V in cold 1 × MOPS buffer. The transfer to the PVDF membrane and the rest of the western blotting protocol was the same as in Section 4.11.
4.14 Intact mass spectrometry
Measurements were performed on the AB SCIEX 5800 TOF/TOF system (Applied Biosystems, California, USA). The protein concentration used was 0.8 mg/mL. A total of 1 μL of this sample was then mixed with 1 μL of a matrix solution of 5 mg/mL α-cyano-4-hydroxycinnamic acid in acetonitrile/water (1:1, v:v), 0.3% TFA on the sample plate. The sample was air dried for 10 min at room temperature. The measurements took place in positive linear mode (measuring range 20,000–100,000 Da). The graphical analysis of the spectra was performed using Data Explorer® software version 4.1 (Applied Biosystems, California, USA).
4.15 Preparation of samples for mass spectrometry
Four replicates of the following samples were run on a 12% home-made SDS denaturing gel for 1 h 20 min on 180 V: CaMKIIL308BzF treated with UV, untreated CaMKIIL308BzF, CaMKIIL308BzF incubated with Ca2+:Calmodulin and then treated with UV, untreated CaMKIIL308BzF incubated with Ca2+:Calmodulin. Both Calmodulin and CaMKIIL308BzF concentrations were set to 8 μM and Ca2+ to 2 mM in CaMKII size exclusion buffer (25 mM Tris pH 8.0, 250 mM NaCl, 1% Glycerol) supplemented with 1 mM TCEP. After electrophoresis, the gels were stained with Coomassie dye, and rinsed with water, and CaMKIIL308BzF bands were excised using a clean scalpel. Gel slices were then subjected to in-gel digestion.
4.16 In-gel digestion of mass spec samples
Excised gel slices were preserved in 50 mM Triethylammonium bicarbonate buffer (TEAB, Sigma-Aldrich). The gel sections were fragmented into smaller pieces and subjected to sequential washing with 50% Acetonitrile (ACN, Fisher Chemical) and 100% ACN buffers. Following alkylation with 50 mM chloroacetamide (CAA, Sigma-Aldrich), the gel pieces underwent an overnight digestion with a 1:40 (w/w) ratio of Lysyl endopeptidase C (FUJIFILM Wako Chemicals) and a 1:20 (w/w) ratio of Trypsin (Serva). The digestion buffer including tryptic peptides was subsequently transferred to new tubes. Gel pieces were then washed twice with 50% ACN to complete the peptide extraction process and transferred to new tubes. Collected samples were subsequently desiccated.
4.17 LC–MS/MS analysis
The desiccated samples were re-suspended with 20 μL of 0.1% Formic Acid (FA, Biosolve Chimie SARL). The LC–MS/MS analysis was performed on the UHPLC Vanquish Neo cooperated with Orbitrap Exploris 480 from Thermo Fisher Scientific. For each sample, 0.25 μL was loaded and fractionated on a 50 cm analytical column (in-house packed with Poroshell 120 EC-C18, 1.7 μm, Agilent Technologies) with reverse-phase gradient. The mobile phase buffer A was composed of 0.1% Formic Acid in ddH2O and mobile phase buffer B was composed of 0.1% Formic Acid in 80% Acetonitrile (FA, Biosolve Chimie SARL). The gradient started at 4% buffer B then linearly increased to 22.5% in 50 min, followed by a linear increase to 45% buffer B in the following 17 min. During the next 0.5 min, the mobile phase was raised to 99% buffer B and washed for another 12 min. The flow was pumped with 0.25 μL/min.
Analytes were sprayed with nano-electrospray ion source and then guided into Orbitrap Exploris 480. Samples were analyzed under data-dependent mode with positive charge +2 to +4. The orbitrap resolution was set to 120,000 with a scan range (m/z) 375–1200 and 300% normalized automatic gain control target. The peptide dissociation was executed with HCD collision energy 30% with resolution 15,000, isolation window (m/z) 1.6. Ions were selected with criteria 1.0e4 intensity threshold coupled with 20 s exclusion duration.
4.18 Mass spec data analysis
The acquired raw data were subjected to analysis using MaxQuant 2.4.2.0, searching against the CaMKIIL308BzF and Calmodulin sequences for the database. The following modifications were considered as variable modifications: Oxidation (M), Acetylation (N-terminus), and Carbamidomethylation (C). These modifications were considered as variable modifications for the analysis. FTMS MS/MS tolerance was set to 20 ppm. Both PSM FDR and Protein FDR were set to 0.01. The ratio-modified: base was used for ANOVA and Student t-test analysis.
AUTHOR CONTRIBUTIONS
Iva Lučić: Conceptualization (equal); data curation (equal); formal analysis (equal); funding acquisition (supporting); investigation (equal); methodology (equal); project administration (equal); validation (equal); visualization (equal); writing – original draft (equal); writing – review and editing (equal). Pin-Lian Jiang: Investigation (supporting); visualization (equal); writing – review and editing (supporting). Andreas Franz: Conceptualization (supporting); investigation (supporting); methodology (supporting); writing – review and editing (supporting). Yuval Bursztyn: Investigation (supporting). Fan Liu: Formal analysis (supporting); investigation (supporting); supervision (equal). Andrew J. R. Plested: Conceptualization (equal); data curation (equal); formal analysis (equal); funding acquisition (equal); methodology (equal); project administration (equal); resources (equal); supervision (equal); validation (equal); visualization (equal); writing – original draft (equal); writing – review and editing (equal).
ACKNOWLEDGMENTS
We are grateful to Alejandro Castro Scalise for purifying Calmodulin, Heike Nikolenko (FMP-Berlin) for intact mass determinations, and Jörg Malsam (BzH, Heidelberg, Germany) for the assistance with expression of BzF-containing proteins in E. coli. Iva Lučić was recipient of a Marie Curie Incoming International Fellowship (798696) and “Wiedereinstiegsstipendium” from the Leibniz FMP. This work was funded by the DFG TRR 186 (Project A07, Projektnummer 278001972 to Andrew J. R. Plested), a DFG Heisenberg Professorship (to Andrew J. R. Plested, Projektnummern 323514590 & 446182550), and the European Research Council (ERC-STG no. 949184 to Fan Liu). Andreas Franz was recipient of a Boehringer Ingelheim PhD Fellowship. Molecular graphics and analyses performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
All relevant data are provided in the paper.