Zero-shot mutation effect prediction on protein stability and function using RoseTTAFold
Review Editor: Nir Ben-Tal
Abstract
Predicting the effects of mutations on protein function and stability is an outstanding challenge. Here, we assess the performance of a variant of RoseTTAFold jointly trained for sequence and structure recovery, RFjoint, for mutation effect prediction. Without any further training, we achieve comparable accuracy in predicting mutation effects for a diverse set of protein families using RFjoint to both another zero-shot model (MSA Transformer) and a model that requires specific training on a particular protein family for mutation effect prediction (DeepSequence). Thus, although the architecture of RFjoint was developed to address the protein design problem of scaffolding functional motifs, RFjoint acquired an understanding of the mutational landscapes of proteins during model training that is equivalent to that of recently developed large protein language models. The ability to simultaneously reason over protein structure and sequence could enable even more precise mutation effect predictions following supervised training on the task. These results suggest that RFjoint has a quite broad understanding of protein sequence-structure landscapes, and can be viewed as a joint model for protein sequence and structure which could be broadly useful for protein modeling.
1 INTRODUCTION
Accurate prediction of single-point mutation effects using sequence information alone would help relate observed sequence polymorphisms to human disease (Hopf et al., 2017; Shin et al., 2021) and contribute to the design of proteins with higher functional activities. Deep learning methods have recently shown considerable promise for mutation effect prediction. DeepSequence (Riesselman et al., 2018), a probabilistic model for sequence families, obtained high accuracy in mutation effect prediction using latent variables for capturing higher-order interactions between residues in proteins through training on multiple sequence alignments (MSAs) for the target protein of interest. Large protein language models trained on MSAs (MSA Transformer) (Rao et al., 2021) or single sequences (Meier et al., 2021) also perform well at mutation effect prediction using an unsupervised or zero-shot approach. These language models have the advantage over DeepSequence of not requiring specific training on the protein family of interest.
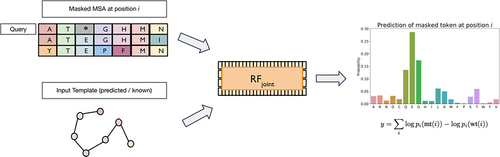
RoseTTAFold was originally developed for protein structure prediction (Baek et al., 2021) and more recently RoseTTAFold Joint (RFjoint) was further trained to solve protein “inpainting” problems (Wang et al., 2022). During the inpainting process using the specifically trained RoseTTAFold network, a pass through the network starts from the functional site and fills in missing sequence and structure, resulting in the creation of a complete and viable protein scaffold. Included in RFjoint training was a masked MSA token recovery task for sequence prediction: predicting the correct amino acid sequence at specific masked positions within the alignment.
To assess RFjoint's understanding of protein mutational landscapes, we set out to investigate whether it could predict experimental mutational data from published deep mutational scanning (DMS) sets (Starita & Fields, 2015) with no further training (i.e., using a “zero-shot” approach). We compared the performance of RoseTTAFold Joint on this task to that of MSA Transformer and DeepSequence. All three are MSA-based methods, RFJoint and MSA Transformer require no further training, while DeepSequence is trained on data from the family of interest. While not developed specifically for this task, we found that the performance in predicting the effects of single mutations on a set of diverse proteins was slightly better for RFjoint than MSA Transformer and comparable to the specifically trained DeepSequence.
2 RESULTS
RFjoint was evaluated on a set of 38 deep mutational scans curated by Riesselman et al. (2018). (The original dataset consisted of 42, we excluded the tRNA (TRNA_YEAST), the toxin–antitoxin complex (PARE_PARD), HIS7_YEAST_Kondrashov2017 and the PABP-doubles datasets to focus on single mutations made to monomeric proteins.) Each of the mutational scans recorded a different protein function with varying measurements. Given that only 2 out of the 38 DMS datasets pertain specifically to stability, the evidence for the stability change prediction is weaker compared to that for the functional effect prediction. Each dataset was treated as a separate prediction task, and each variant was scored individually. For each target protein, we generated MSAs using iterative sequence search against the UniClust30 database as described in Baek et al. (2021) and used it for both RFjoint and MSA Transformer predictions. For RFjoint, the variants were scored by masking out the mutation site in the query sequence in the MSA, and the MSA token recovery head was used to predict the distribution over the masked position. The predicted effect of the mutation was calculated as the log odds ratio of the mutant amino acid and the wild-type amino acid (Figure 1). The performance on each dataset was assessed based on the spearman correlation of the predictions to the observed experimental values. For DeepSequence, we compared the results of MSA Transformer and RFjoint to the published spearman rho values (Riesselman et al., 2018), which are from an ensemble of models trained on a different set of MSAs than those used for MSA Transformer or RFjoint for each target protein.
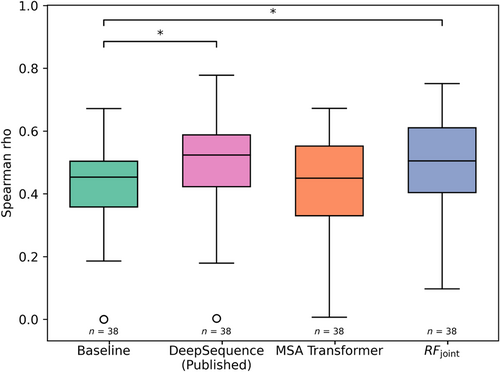
We found that RFjoint predicts mutational effects considerably better than a baseline calculated as the log odds ratio of the frequency of the mutant amino acid and of the wild-type amino acid in the MSA (Figure 2). RFjoint also slightly outperformed MSA Transformer and is comparable to the protein family-specific DeepSequence (Figure 2). RFjoint has the advantage in principle over the purely sequence-based models of also being able to utilize structural template information, but we did not observe a significant improvement with incorporation of template structure information (Supplementary Figure S1; this may be in part because RoseTTAFold generates 3D models from MSA with reasonable accuracy). We also found little dependency of prediction accuracy on MSA depth (Supplementary Figure S2).
3 DISCUSSION
We find that the RoseTTAFold network developed originally for structure prediction and then extended to protein design, is also able to predict the effect of single mutations with quite a high accuracy. DeepSequence has a slightly higher average spearman rho correlation than RFjoint but requires training for each protein family individually. Just as large protein language models, like MSA Transformer, provide general models of protein sequence, RoseTTAFold Joint may be viewed as a general joint model of protein sequence and structure. With further directed training, it should be possible to further improve mutation effect prediction performance by better-utilizing protein structural information, which can be readily input into RoseTTAFold Joint but not into pure sequence-based models, and by fine-tuning specifically on the mutant prediction task. The ability of RoseTTAFold to function as a joint model of protein sequence and structure, incorporating any available protein sequence and structure information could prove useful in applications beyond protein design and mutation effect prediction.
4 MATERIALS AND METHODS
4.1 Deep mutational scanning datasets
RoseTTAFold was evaluated on a subset of 38 deep mutational scans collected by Riesselman et al. (2018). The proteins evaluated perform a wide range of functions and the experimental measures performed are different for each protein. We treat each DMS dataset as a separate prediction task. Performance on each task is evaluated by spearman rho correlations of the calculated (baseline), published (DeepSequence), or predicted (RFjoint and MSA Transformer) scores to the experimental values.
4.2 MSA generation
The same MSA inputs are used for both RoseTTAFold Joint and MSA Transformer at inference time. The protocol for generating MSAs is adopted from RoseTTAFold (Baek et al., 2021), where for each protein, sequences are found by iterative search against UniRef30 (Mirdita et al., 2017) and BFD (Steinegger, Mirdita, & Söding, 2019) using HHblits (Steinegger, Meier, et al., 2019). Sequences are then filtered at 90% sequence identity cutoff. The E-value cutoff for sequence search is gradually relaxed (from 1e-10 to 1e-3) until the generated MSA has at least 2000 sequences with 75% coverage or 5000 sequences with 50% coverage. For the proteins that failed to get 5000 sequences (with E-value of 1e-3 and 50% sequence coverage cutoff), as many sequences as the protocol can find are used as an input MSA.
4.3 Non-ML baseline setup
4.4 RFjoint inference setup
4.5 MSA transformer inference setup
We used the published MSA Transformer (Meier et al., 2021; Rao et al., 2021) loaded with pre-trained weights (annotated as esm_msa1b_t12_100M_UR50S on the public ESM github). The default arguments were used, where 400 sequences were randomly sampled from the MSA for inference. We used the masked marginals scoring strategy for scoring mutants from MSA Transformer, which is done by introducing masks at the mutated positions and computing the score for a mutation by considering its probability relative to the wildtype amino acid (Meier et al., 2021). This is similar to the setup that we used for predicting the effect of a mutation through RFjoint (Equation (2)).
AUTHOR CONTRIBUTIONS
Sanaa Mansoor: Conceptualization; Investigation; Methodology; Validation; Visualization; Data curation; Writing—original draft; Writing—review & editing; Formal analysis. Minkyung Baek: Conceptualization; Methodology; Writing—review & editing; Writing—original draft; Formal analysis. David Juergens: Methodology; Writing—original draft. Joseph L. Watson: Methodology; Writing—original draft. David Baker: Conceptualization; Methodology; Validation; Supervision; Writing—original draft.
ACKNOWLEDGMENTS
We would like to thank Justas Dauparas, Ivan Anishchanka, Doug Tischer, Hahnbeom Park, Sergey Ovchinnikov, and Eric Horvitz for helpful comments and suggestions.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Inference code for predicting the effect of single mutations on protein function or stability through this pipeline is available here: https://github.com/RosettaCommons/RFDesign/tree/main/inpainting. All input data (target MSAs, structural templates), and experimental and predicted values of all methods compared are available on Zenodo at this link: https://doi.org/10.5281/zenodo.8106250.