Hydrophobicity and molecular mass-based separation method for autoantibody discovery from mammalian total cellular proteins
Abstract
Serum autoantibody profiles are unique to individuals and reflect the level and history of autoimmunity and tumor immunity. The identification of autoantibody biomarkers is critical for the development of immune monitoring systems for immune-related disorders. Here, we present a practical method for large-scale autoantibody discovery using total cellular proteins from cultured mammalian cells. We found that nucleic acid-free and fully denatured water-soluble total cellular proteins from mammalian cells were superior, allowing precise separation by reversed-phase HPLC after preparing a large set of homogeneous total cellular proteins. After separating the proteins based on hydrophobicity, the fractionated samples were subjected to molecular mass analysis using conventional SDS-PAGE. The resulting two-dimensional gel electrophoresis was successfully employed for immune blotting and LC–MS/MS analysis. All procedures, including TRIzol-based total cellular protein extraction, solubilization of denatured proteins, reversed-phase HPLC separation, and SDS-PAGE, were highly reproducible and easily scalable. We propose this novel two-dimensional gel electrophoresis system as an alternative proteomics-based methodology suitable for large-scale autoantibody discovery.
Abbreviations
-
- 2-DE
-
- two-dimensional gel electrophoresis
-
- GdnHCl
-
- guanidine hydrochloride
-
- HYD
-
- hydrophobicity
-
- MM
-
- molecular mass
-
- MS
-
- mass spectrometry
-
- pI
-
- isoelectric point
-
- TCL
-
- total cell lysate
-
- TCP
-
- total cellular protein
1 INTRODUCTION
The presence of autoantibodies in the blood is thought to occur because of a breakdown in self-tolerance. Some recognize conformational epitopes on extracellular proteins that may disrupt the normal function of critical proteins or pathways (Burbelo et al., 2021; Wang et al., 2015). However, various autoantibodies are frequently observed in healthy individuals (Ludwig et al., 2017; Nagele et al., 2013; Neiman et al., 2019; Shome et al., 2022), and the link between the antigenicity and pathogenicity of many autoantibodies is less clear. For example, autoantibodies against intracellular autoantigens do not directly contribute to pathogenesis because they cannot bind to antigens in living cells. Instead, serum autoantibodies potentially reflect the level and history of inflammatory reactions, as the released intracellular proteins can stimulate humoral immunity. Thus, autoantibodies are potential biomarkers of abnormal autoimmune reactions triggered by inflammation (Burbelo et al., 2021; Wang et al., 2015).
Autoantibodies are promising biomarkers that reflect the level of anti-tumor immune reactions. When the immune system eliminates cancer cells, intracellular cancer-specific proteins that leak from dead cancer cells stimulate humoral immune responses, increasing the autoantibodies against cancer-specific antigens (Brossart, 2020; Gnjatic et al., 2010; Gottschalk et al., 2013; Kobayashi et al., 2020; Nesslinger et al., 2010; Ohue et al., 2019; Ohue, Kurose, et al., 2014; Ohue, Wada, et al., 2014; Zaenker et al., 2016). Anti-NY-ESO-1 and anti-XAGE-1b, both categorized as cancer-testis antigens (CTAs), have been reported as potential predictive biomarkers for good response and prolonged survival with anti-PD-1 monotherapy in non-small cell lung cancer (NSCLC) (Ohue et al., 2019; Sakai et al., 2021). The recently developed immune checkpoint inhibitors (ICIs) represent a significant breakthrough in cancer immunotherapy for various types of cancer (Liebl & Hofmann, 2019; Shiravand et al., 2022). However, the response rate to ICIs is still only 20%–40%, highlighting the need to develop predictive biomarkers.
Many clinical studies have shown that individual differences in clinical outcomes of ICI therapy are closely related to individual differences in the tumor microenvironment (TME) and immune status (Binnewies et al., 2018). For instance, immune cell-enriched tumors classified as immune-favorable TME subtypes benefit the most from ICI therapy (Bagaev et al., 2021). Tumor profiling by RNA sequencing of tumor biopsies allows reliable evaluation of the immune status of the tumor; however, repetitive invasive tumor biopsies are unavailable at clinical sites. Thus, autoantibody biomarkers in the peripheral blood are expected to be complementary diagnostic tools. The higher stability of IgG autoantibodies in peripheral blood is also an advantage for diagnostic use in clinical sites (Yang et al., 2022).
Recent immuno-oncological studies also suggest that autoantibody biomarkers have the potential to reflect the TME. Immune cells in the TME can form tertiary lymphoid structures (TLSs) through long-lasting exposure to inflammatory signals mediated by chemokines and cytokines (Sautès-Fridman et al., 2019). TLSs, often observed in the TME, correlate with improved prognosis in cancer immunotherapy because of the influential local generation of autoreactive T and B cells and autoantibodies that activate the cancer-immunity cycle (Mazor et al., 2022). Based on this immuno-oncological aspect, patients who exhibit increased levels of various autoantibodies in peripheral blood are likely to have a more potent prognosis by activating cancer immunity. Although an autoantibody-based prediction system for cancer immunotherapy is not yet clinically applicable, a simple blood test-based evaluation can potentially enable precise cancer therapy.
One issue in developing diagnostic tools for cancer immunology or autoimmune diseases using autoantibody biomarkers is the significant interindividual variability in autoantibody profiles. Therefore, it is essential to develop comprehensive autoantibody panels to detect a wide range of autoantibodies. Simultaneously, the selection of disease-specific autoantibody biomarkers that are frequently detectable in patient serum is required to lower assay costs. Serological proteome analysis (SERPA) of various clinical specimens is crucial for achieving this goal (Beutgen et al., 2019).
Conventional approaches to SERPA widely employ two-dimensional gel electrophoresis (2-DE) (Klose, 1975; O'Farrell, 1975), which separates proteins according to their isoelectric point (pI) and molecular mass (MM), followed by western blotting using patient serum. Although the conventional 2-DE approach has yielded many successful results (Bandow et al., 2008), achieving highly reproducible results at the laboratory level is often challenging because of multistep gel-based processing and the need for careful sample preparation. A protein microarray technique, which is a high-throughput method for the simultaneous analysis of multiple samples (Beutgen et al., 2019; Ganesan et al., 2016; Morishita et al., 2019; Yang et al., 2016), can also be helpful; however, it may incur a high cost if it requires the study of many clinical samples.
Highly reproducible protein separation using MM allows the use of commercially available high-quality polyacrylamide gels. However, a highly complex mixture of whole-cell lysates makes discrimination between bands challenging. This protein complexity can be partially diminished using the detergent-dependent solubility of cellular proteins and separating them into three distinct fractions: cytoplasmic, membrane/organelle, and nuclear/cytoskeletal proteins (Lenstra & Bloemendal, 1983). Nucleic acids, which are always present in cell lysates, are also known to affect protein separation; thus, enzymatic digestion or sonication is widely employed in sample preparation procedures (Wu, 2006). Removal of nucleic acids from total cellular proteins (TCP) under native conditions is intrinsically challenging. In a previous study, we discovered that fully denatured mammalian total cell protein mixtures show unusually high solubility in nucleic acid-free pure water (Futami et al., 2014). Although the detailed mechanism is unclear, mammalian intracellular protein mixtures possess aggregation-deficient biophysical properties as they are rich in intrinsically disordered regions and favor highly flexible hydration in pure water rather than aggregate formation. This study proposes a novel and highly reproducible 2-DE technique for separating TCP by hydrophobicity (HYD) and MM using TCP solubilization techniques and reversed-phase HPLC.
2 RESULTS
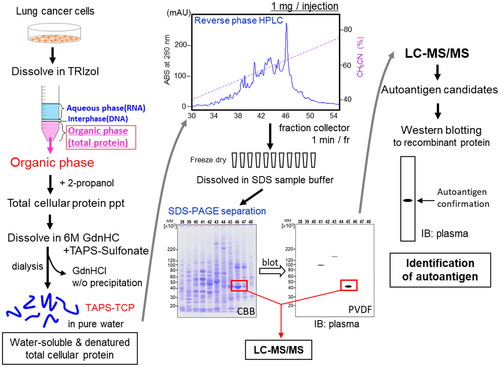
2.1 Schematic workflow of autoantibody biomarker discovery by TRIzol-based TCP extraction, reversed-phase HPLC and 2-D PAGE
Autoantibody discovery from patient serum or plasma requires an antibody–antigen reaction. In this study, antigens for TCP were obtained from cultured cell lines (Figure 1). This procedure could also be applied to tissue-derived samples. TRIzol-based protein extraction is a simple and scalable method that minimizes endogenous protease digestion (Futami et al., 2014). The TRIzol extraction procedure quantitatively extracts TCP with complete removal of nucleic acid contamination. After dissolving the extracted proteins in 6 M guanidium hydrochloride (GdnHCl), all cysteine residues were modified via the alkyl disulfide reaction, which enhanced protein solubility by introducing positive charges by TAPS-sulfonate (Futami et al., 2015; Futami et al., 2017; Kimura et al., 2020; Miyamoto et al., 2022; Murata et al., 2006). Highly water-soluble TCP was recovered by dialyzing against pure water without any visible precipitates. Reversed-phase HPLC was used to separate proteins according to their hydrophobicity. The fractionated samples were lyophilized and dissolved in SDS-sample buffer. These scalable fractionated samples allowed multiple homogeneous samples to be subjected to SDS-PAGE, enabling their long-term storage under freezing conditions. Notably, this procedure allows the preparation of homogenous multiple polyvinylidene difluoride (PVDF) blots using the above-fractionated samples on a quality-certified commercially available polyacrylamide gel. The subsequent autoantibody discovery procedure was the same as that generally employed in SERPA, including western blotting using patient serum or plasma, in-gel digestion of the targeted protein bands, LC–MS/MS analysis to identify the target antigens, and confirmation of autoantibody reactivity using recombinant proteins.
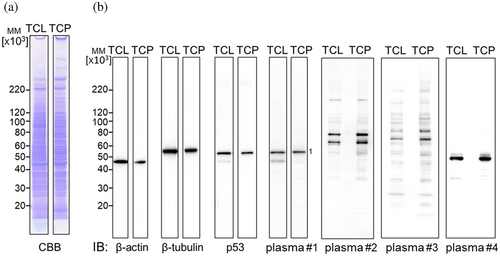
2.2 Quantitative extraction of TCP using TRIzol
The quality of TCP extracted from cultured cells by TRIzol was validated by comparing it with the total cellular lysate (TCL) obtained by directly lysing the cells in lysis buffer. The protein contents of both TCL and S-cationized TCP by TAPS-sulfonate (TAPS-TCP) were found to be comparable by Coomassie Brilliant Blue (CBB) staining (Figure 2a), indicating the quantitative extraction of total cellular protein using TRIzol. The western blotting band intensities of the internal controls, β-actin, and β-tubulin were also comparable; thus, both samples contained comparable intracellular proteins (Figure 2b). When the diluted plasma from patient #1 with lung cancer was reacted with the PVDF blots, clear bands of approximately 50 kDa (band-1) were detected. Identical reaction patterns with anti-p53 specific antibodies confirmed the presence of an autoantibody-reactive band. The immunoblotting patterns of patients 2, 3, and 4 were comparable in both TCL and TCP, indicating that TCP includes total cellular proteins without losing them during the TRIzol extraction and dialysis steps. Interestingly, the native TCL included a degraded band of approximately 45 kDa, which was not observed for TAPS-TCP. This change may reflect the endogenous protease activity (Figure 2b). The strong detergent TRIzol completely denatured the endogenous protease, whereas the native TCL may have degraded the p53 protein due to the remaining endogenous protease during lysate preparation.
2.3 Separation of TCP by reversed-phase HPLC
We successfully separated water-soluble TCP mixtures that were nucleic acid-free and fully denatured by hydrophobicity. TAPS-TCP, which increased the hydrophilicity by S-cationization (Futami et al., 2015; Futami et al., 2017; Miyamoto et al., 2022; Murata et al., 2006), shifted the chromatographic profile to an earlier elution than nonconjugated TCP (Figure 3a). Total cellular proteins were distributed from 30 to 54 min of elution, with highly discriminated protein bands. The reversible disulfide bonds in TAPS-TCP-conjugated 3-(trimethylammonium)propyl moieties result in proteins migrating as nonconjugated bands on SDS-PAGE under reducing conditions. The native conditions of TCL with nucleic acids also allowed its separation by reverse-phase HPLC (Figure 3b). Extensively degraded nucleic acid fractions were observed around pass-through fractions. Because the elution time for proteins was lesser than that for the nondenaturing detergent Nonidet® P40, almost all protein UV absorption was monitored (Figure 3b). The elution patterns of denatured TAPS-TCP and native TCL were quite different. SDS-PAGE analysis revealed broader protein bands (19 min, from 33 to 52 min, Figure 3a) for TAPS-TCP and a narrower distribution (15 min, 36 to 51 min, Figure 3b) for TCL. It is concerning that the separation of hydrophobic proteins by reversed-phase HPLC can sometimes result in a ghost peak because of residual protein sample adsorption. However, no peaks were observed in the second elution cycle (Figure S1).
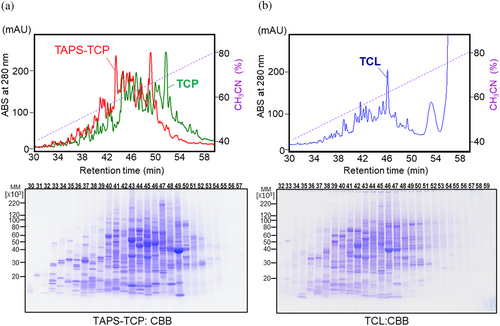
This methodology has been effectively extended to hydrophobic integral membrane proteins (Figure S2). The integral membrane proteins of coxsackie and adenovirus receptor (CAR) (Ortiz-Zapater et al., 2017), endogenously expressed in HeLa cells, were solubilized as TAPS-TCP and subsequently separated by reversed-phase HPLC. This separation yielded distinct protein entities, visualized through HYD-MM 2-DE blots employing specific anti-CAR antibodies. Furthermore, a highly effective elution buffer designed for hydrophobic protein separation in a reversed-phase HPLC column, consisting of a mixture of acetonitrile and 2-propanol (2:1) (Tarr & Crabb, 1983), showed no detectable peaks during the second elution cycle. Thus, this methodology enables proteomics even for hydrophobic membrane proteins.
2.4 Demonstration of controllable resolution and validation of reproducibility for 2-D protein separation
We successfully validated the HYD-MM 2-DE system, which utilizes TAPS-TCP from NCI-H1975 cell culture, as it has a controllable resolution for protein separation (Figure 4a) and demonstrated high reproducibility in sample preparations (Figure 4b,c).
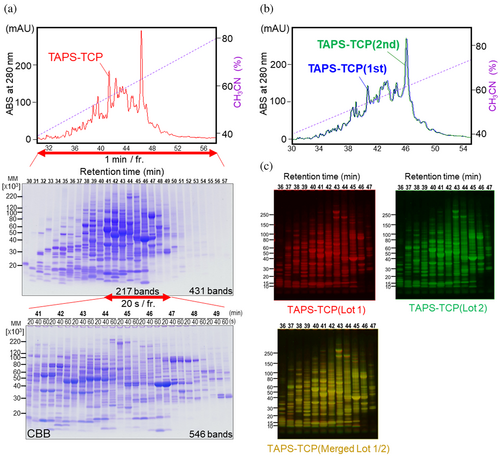
A total of 431 discriminated bands were identified as detectable proteins at elution times ranging from 30 to 57 min when fractionated for 1 min each (Figure 4a, middle). The band resolution was controlled by adjusting the fractionation time. Highly condensed bands, consisting of 217 bands in the elution time range of 41–49 min, could be expanded to 546 bands when the fractionation time was reduced from 60 to 20 s. This demonstrated that the resolution of the discriminated protein bands could be adjusted as required.
The reproducibility of the 2-D separation is also critical for proteomic analysis. The inter-injection (1 mg of protein each) reproducibility of TAPS-TCP on reversed-phase HPLC confirmed a similar chromatographic trace pattern between the first and second injections (Figure 4b). Thus, the repetitive separation of TAPS-TCP by HPLC resulted in homogenously fractionated proteins. This allows for scalable proteomic analysis that is favorable for a large set of autoantibody screening using identical 2-D separated PVDF blots and gels. Inter-sample reproducibility was also demonstrated using TAPS-TCP prepared from different cell culture lots. Although the HPLC chromatographic trace pattern showed slight changes depending on the preparation of the cell culture lots (Figure S3), the HYD-MM 2-DE profiles were comparable, thus establishing a highly reproducible procedure (Figure 4c).
2.5 Demonstration of SERPA using HYD-MM 2-DE
The optimized HYD-MM 2-DE gel of total cellular proteins from NCI-H1975 cells was used to identify target antigens that reacted with autoantibodies in the plasma of patient #5 with lung cancer using the SERPA method. As shown in Figure 5a,b, HYD-MM 2-DE gels and blots were prepared on commercial minigels. The immunoreactive bands at approximately 55 kDa on the 1-D gel separated into four distinct bands on the HYD-MM 2-DE gel (Figure 5b). Proteomic analysis of each antigen revealed vimentin (band-2), keratin-7 (band-3), enolase-1 (band-4), and keratin-8 (band-5) as the target proteins. The three specific antibodies from our stock exhibited specific immune reactions against the corresponding antigens (Figure 5c–e).
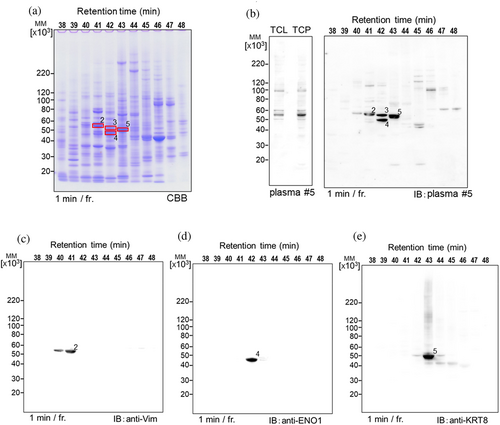
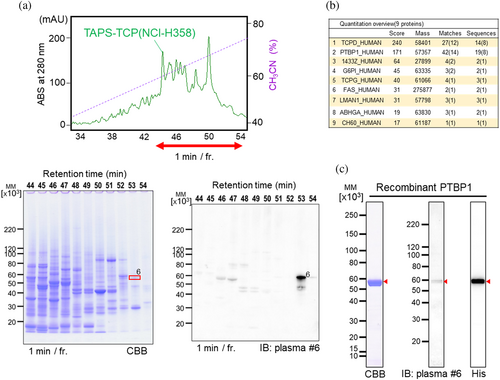
Another cell line, NCI-H358 derived TAPS-TCP, was used for autoantibody screening (Figure 6a). After excising the target band corresponding to the immunoreactive band on the PVDF blots and performing in-gel digestion, candidate target antigens were identified using MASCOT search analysis (Figure 6b). The critical criterion for selecting target antigens among these candidates is data mining to match autoimmune or cancer-related antigens. The recombinant protein expressed as a HisTag-fused protein facilitated the purification and detection of the target antigen, which was then checked for an immune reaction against the autoantibody-targeted antigen (Figure 6c). Using this procedure, we identified 26 autoantigens from 14 patients with lung cancer (Figures 2, 5 and 6, S4).
2.6 Autoantibody detection via indirect immunofluorescence cytochemistry
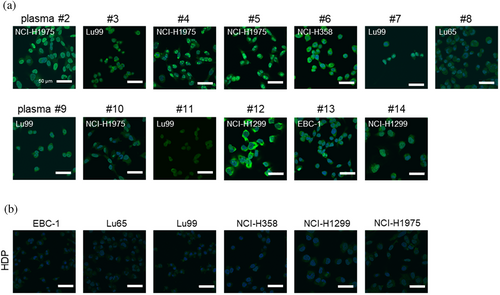
Visualization of autoantibodies present in the plasma of patients with lung cancer was accomplished using indirect immunofluorescence cytochemistry against lung cancer-derived cell lines (Figure 7). Autoantibody reactions against the cell lines varied between individuals, and the cells that exhibited the higher reactions were identified by 1-D gel western blotting (Figure S5). Figure 7a shows clear autoantibody reactions against each cell line visualized in paraformaldehyde-fixed and permeabilized cells. The topographic distribution of the immunofluorescence patterns also varied among individuals, suggesting diversity and individual differences in immune responses. Notably, healthy donor pooled (HDP) serum from 10 healthy donors showed no clear autoantibody reactions (Figure 7b). These results demonstrate that autoantibodies in patients with lung cancer react with lung cancer-derived cell-derived proteins, and proteomic analysis revealed some targeted antigens.
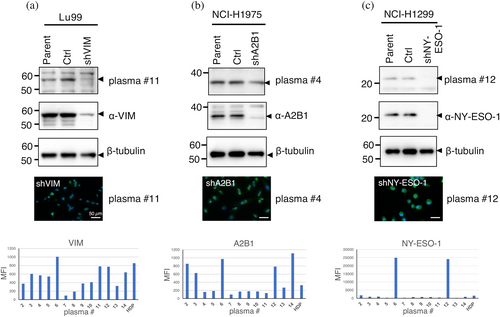
Three of the identified autoantigens were confirmed to exhibit specific binding in the knockdown experiments (Figure 8). Endogenous proteins in each cell were found to react with autoantibodies in the plasma on a 1-D gel. In contrast, the target bands were significantly downregulated in knockdown cells. Interestingly, the autoantibody reaction in indirect immunofluorescence cytochemistry showed no significant reduction in fluorescence intensity. These results also support the conclusion that the autoantibodies present in peripheral blood are composed of multiple autoantibodies directed against different autoantigens. Autoantibody levels were quantified using Luminex beads immobilized with antigens (Futami et al., 2015; Miyamoto et al., 2022) which confirmed the heterogeneity of autoantibody levels among individuals. As all identified antigens were selected based on their binding to autoantibodies on PVDF blots, it can be inferred that autoantibodies specifically recognize linear epitopes on the denatured form of antigens. The water-soluble and denatured forms of the antigens immobilized on the beads through S-cationization demonstrated robust reactivity with autoantibodies, making them suitable for quantitative assays.
3 DISCUSSION
Highly discriminated separation of 2-D gels is critical for proteomic studies. Traditional reversed-phase HPLC is an accessible and highly reproducible system for separating proteins based on hydrophobicity. TCL (directly lysed cells in lysis buffer) and TCP (proteins extracted using TRIzol) were used as sources in this proteomics study. Fully denatured and water-soluble TCP exposed hydrophobic residues, resulting in broader separation on reversed-phase HPLC (Figure 3). Notably, this methodology only applies to proteomic studies of mammalian total cellular proteins because these TCP extraction methods cannot fully solubilize prokaryotic total cellular proteins (Futami et al., 2014). Nucleic acid-free and denatured mammalian TCP mixtures exhibit high solubility in pure water. S-cationization by TAPS-sulfonate enhances the water solubility of the conjugated proteins. This effect is likely attributable to the preferential hydration of enriched, intrinsically disordered proteins, which interferes with hydrophobic interactions. Although the precise mechanism underlying this unique solubility remains of biophysical interest, water solubility is a crucial advantage for separating proteins by hydrophobicity in reverse-phase HPLC for hydrophobicity-mediated preparation of 2-D gel electrophoresis (HYD-MM 2-DE) preparations.
Mammalian TCP is a robust sample suitable for large-scale proteomic analysis because it can pass through 0.45 μm pore size syringe filters and can be stored long-term at 4°C without aggregation and proteolytic degradation. The reversible S-cationization of TCP by TAPS-sulfonate improved the physicochemical properties of the sample by increasing the hydrophilicity of denatured proteins in reversed-phase HPLC and provided higher resolution for the separation of total cellular proteins (Figure 3a). Using native TCL remains a valuable alternative for generating distinct patterns within HYD-MM 2-DE gels (Figure 3b). While the protein conformations within native TCL can be disrupted under acidic conditions, such as 0.1% HCl, the initial separation pattern obtained by reversed-phase HPLC was markedly different from that obtained with fully denatured TCP or TAPS-TCP. This dissimilarity may be due to the partial persistence of residual intact protein conformation, even under acidic conditions. A similar two-dimensional microchip array, combining the first pI-focused fractions from the cell lysate and the second separation by a reversed-phase HPLC, has also been proposed as a high-resolution proteomics tool (Yan et al., 2003; Yu et al., 2019). Thus, alternative 2-DE separation systems suitable for the analysis should be selected for each experiment.
Autoantibodies are highly stable proteins present in the peripheral blood that serve as crucial biomarkers for reflecting autoimmune reactions and are potent indicators for predicting or monitoring the clinical outcomes of cancer immunotherapy. Immune-related adverse events (irAEs) are often observed with ICI therapy because of an accelerated immune response (Chen et al., 2015). A recent immuno-oncology study showed that irAEs associated with ICI therapy are closely associated with a better clinical prognosis (Freeman-Keller et al., 2016; Zhou et al., 2020). Thus, to develop precision medicines, it is crucial to predict the risk of irAEs and explore the threshold of autoantibody profiles that induce irAEs in ICI therapy, especially in intractable cancers. Real-time monitoring of the immune status is a powerful tool for realizing precision medicine. Autoantibody biomarkers, which are closely related to autoimmune and anti-tumor immune reactions, will be one of the critical molecules with easily handled in clinical settings. The selection of a set of autoantigen panels is crucial for better diagnosis and prognostic prediction. The autoantibody profile's inter-individual variability must be considered for the development of an immune-monitoring platform. Figure 8 shows autoantibody profiles and levels within lung cancer patients showed inter-individual variability. Therefore, the autoantibody assay platform requires a semi-comprehensive antigen array with a validated quantitative assay system, which is our ongoing study. The immune monitoring platform's selection of disease-related autoantibodies is an essential issue. In this study, most of the 26 autoantigens identified from patients with lung cancer were closely related to autoimmune diseases rather than to cancer. This finding strongly suggests that autoimmunity-related autoantibodies are synergistically induced during the cancer-immunity cycle in each cancer patient. Selecting frequently observed autoantibodies in each disease patient group using SERPA could lead to the development of immune profiling and monitoring tools.
Western blotting-based serological proteomic analysis for autoantibody biomarker screening has limitations when detecting antibodies against conformational epitopes, because it only detects linear epitopes. However, this limitation is less significant in the field of immuneoncology. Analysis of epitopes recognized by autoantibodies against cancer-testis antigens in patients with lung and esophageal cancer revealed that their ability to bind to diverse linear epitopes varies among individuals (Kawabata et al., 2007). Thus, while full-length autoantigens are necessary, polyclonal antibodies are not required to detect their conformation. This inclination of autoantibodies towards linear epitopes is believed to be influenced by autoantigen conformation. Notably, the majority of cancer-testis antigens belong to a class of intrinsically disordered proteins (Ahmadi et al., 2021; Rajagopalan et al., 2011). Although most intrinsically disordered proteins adopt biologically active structures through interactions with binding partners, proteins aberrantly expressed in cancer cells lack such associations, resulting in their aggregation or degradation (Ahmadi et al., 2021). This biophysical attribute potentially contributes to the antigenicity of the humoral immune responses. Consequently, many recombinant autoantigen proteins demonstrate an aggregation propensity. The S-cationization technique is valuable for the preparation of recombinant autoantigens with fully exposed high-quality linear epitopes. Linear epitope-specific autoantibodies can be effectively quantified using an S-cationized antigen-immobilized multiplex bead array (MUSCAT) assay system (Futami et al., 2015; Miyamoto et al., 2022).
This study presents an innovative approach to autoantibody discovery centered on fully denatured protein-based proteomics. This methodology is based on the solubility of denatured proteins in pure water. The expansion of techniques that enable the handling of denatured proteins offers novel insights into the behavior of such proteins and opens avenues for further advancements in protein research.
4 MATERIALS AND METHODS
4.1 Cell culture and patient plasma
Human non-small cell lung cancer-derived EBC-1, Lu65, Lu99, NCI-H358, NCI-H1299, and NCI-H1975 cells (ATCC) and human cervical epithelioid carcinoma (HeLa) cells (ATCC) were cultured in RPMI1640 medium (Wako Chemical, Osaka, Japan) supplemented with 10% FBS in a humidified incubator. HDP from 10 healthy donors (serum from five males and five females, ages 19–49 years) were obtained from Tennessee Blood Services (TN, USA). Plasma samples from patients with lung cancer who provided written informed consent were obtained from the Okayama University Hospital Biobank (Okadai Biobank, Japan), in collaboration with Dr. Kiura K. and Dr. Ohashi K. (Okayama University Hospital) for biomarker screening, which was approved by the ethics committee of Okayama University.
4.2 Preparation of native cell lysate and water-soluble nucleic acid-free denatured total cellular protein mixtures
Subconfluent lung cancer cells cultured in 10 cm dishes were used in this study. The TCL under native conditions was prepared by dissolving cells in lysis buffer (20 mM HEPES-NaOH buffer, pH 7.5, 50 mM NaCl, 2 mM MgSO4, 1% Nonidet® P40) supplemented with a protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan) and ruptured for 12.5 min (250 W, cycle of 20 s ON and 20 s OFF) on ice using a sonicator (Bioruptor UCD-250, CosmoBio, Tokyo, Japan). To digest nucleic acids, 125 unit/mL of Benzonase (Merck Millipore, Burlington, MA, USA) were added and incubated for 30 min at 25°C. Nucleic acid-free, fully denatured, and water-soluble total cellular proteins were prepared using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA) according to previously described methods (Futami et al., 2014). The recovered denatured protein precipitates were solubilized in 6 M GdnHCl containing 0.1 M Tris–HCl (pH 8.5) and reduced with 30 mM dithiothreitol for 2 h at 37°C. S-cationization of all Cys residues in total cellular proteins was prepared by adding 70 mM [3-(trimethylammonium)propyl] methanethiosulphonate (TAPS-sulfonate, Katayama Chemical, Osaka, Japan) and incubating for 1 h at 37°C (Futami et al., 2015; Futami et al., 2017; Kimura et al., 2020; Miyamoto et al., 2022; Murata et al., 2006). Total cellular protein, with or without S-cationization, was adjusted to a protein concentration of 3 mg/mL with 6 M GdnHCl and mixed with a 0.1 volume of acetic acid. After extensive dialysis against pure water at 4°C, water-soluble nucleic acid-free denatured TCP mixtures were recovered without visible precipitation.
4.3 HYD-MM 2-DE
All total cellular proteins from TCL, TCP, and S-cationized TCP (TAPS-TCP) were filtered using 0.45 μm pore size syringe filters (Merck Millipore) before applying for HPLC. TCL protein concentrations were determined using the Bradford protein assay (Bio-Rad Laboratories, Hercules, CA, USA) with BSA as a standard. TCP and TAPS-TCP were determined by measuring the absorbance at 280 nm at 1OD = 1 mg/mL. One milligram of each protein sample was separated on a reversed-phase HPLC column (COSMOSIL Protein-R, 4.6 mm I.D. × 150 mm, Nacalai Tesque) using an acetonitrile linear gradient elution procedure in the presence of 0.1% HCl. The eluted proteins were collected every 60 or 20 s using a fraction collector and lyophilized using a centrifugal evaporator (CVE-200D; Eyela, Tokyo, Japan). Dried samples were then dissolved in SDS-PAGE sample buffer with a reducing reagent (Nacalai Tesque) and subjected to SDS-PAGE analysis on 5%–20% gradient mini-gels (Wako Chemical) or on a wide gel (ATTO, Tokyo, Japan). The fractionated samples were applied at 1/8 volume for the mini gels and 1/4 volume for the wide gels.
4.4 Immunoblot analysis
The following monoclonal (mAb) and polyclonal (pAb) primary antibodies were used for immunoblotting: CAR mAb (D3W3G, Cell Signaling Technology), β-actin mAb (13E5, Cell Signaling Technology, Danvers, MA, USA), β-tubulin mAb (10G10, Wako Chemical), anti-His-tag mAb (OGHis, MBL, Tokyo, Japan), vimentin mAb (D21H3, Cell Signaling Technology), and HNRNPA2B1 pAb (Proteintech, Rosemont, IL, USA). Rabbit polyclonal antibodies against human p53 (Uniport: P04637), alpha-enolase (ENO1, Uniport: P06733), keratin-8 (KRT8, Uniport: P05787), and cancer/testis antigen 1 (NY-ESO-1, Uniport: P78358) were prepared at Cosmo Bio (Tokyo, Japan) by immunizing rabbits with full-length antigens purified using a previously described procedure (Futami et al., 2015; Miyamoto et al., 2022). Target bands were visualized using horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG antibodies (Cell Signaling Technology) with a chemiluminescent reagent (Western Lightning Plus-ECL; PerkinElmer, MA, USA) on an Amersham ImageQuant 800 (GE Healthcare, Chicago, IL, USA).
4.5 Proteomics
After two rounds of sample separation using SDS-PAGE, one gel was subjected to CBB staining and the other was transferred onto PVDF membranes (Immobilon-P, Merck Millipore) using the semi-dry transfer method. The PVDF blots were subsequently blocked using Blocking One (Nacalai Tesque), incubated with human plasma diluted 400-fold in Can Get Signal solution 1 (Toyobo, Osaka, Japan), and probed with horseradish peroxidase-conjugated anti-human IgG (MBL). Western blot results were visualized by adjusting the size of the CBB-stained gel, and the target band was excised under transmitted light. All CBB-stained gel fragments were destained, reduced, and alkylated before tryptic digestion of the proteins. In-gel digestion of samples was performed using 10 ng/μL trypsin-TPCK TREATED (Worthington). The digested protein samples were subjected to HPLC-Chip/QTOF analysis (G6520 and G4240; Agilent). Candidate peptide sequences were analyzed using the MASCOT software (Matrix Science Inc., MA, USA).
4.6 Identification of autoantigens
Putative biomarker proteins were identified using LC–MS/MS and MASCOT search analyses. Full-length and mature forms of Heterogeneous Nuclear Ribonucleoprotein R (HNRNPR), FK506-Binding Protein 4 (FKBP4), Ribosomal Protein L7a (RPL7A), Emerin (EMD), Heterogeneous Nuclear Ribonucleoprotein A2/B1 (HNRNPA2B1), T-Complex Protein 1 (TCP1), Eukaryotic Elongation Factor 2 Kinase (EEF2K), Heat Shock Protein 60 (HSPD1), Lysophosphatidylcholine Acyltransferase 1 (LPCAT1), Alpha-Enolase (ENO1), Keratin 7 (KRT7), Polypyrimidine Tract-Binding Protein 1 (PTBP1), Keratin 8 (KRT8), Peptidyl-Prolyl Cis-Trans Isomerase B (PPIB), Eukaryotic Elongation Factor 1 Gamma (EEF1G), Keratin 19 (KRT19), Plectin (PLEC), Eukaryotic Translation Initiation Factor 3 Subunit A (EIF3A), TAR DNA-Binding Protein 43 (TARDBP), Keratin 18 (KRT18), Vimentin (VIM), Lamin A/C (LMNA), NY-ESO-1/CT6.1, Heterogeneous Nuclear Ribonucleoprotein A1 (HNRNPA1), and FK506-Binding Protein 1A (FKBP1A) were cloned into pET28b vectors (Novagen) to express a His-tag (MGSSHHHHHHSSGLVPRGSH) at the N-terminus, and a StrepTagII (GPGWSHPQFEK) at the C-terminus. Recombinant proteins were expressed in Escherichia coli BL21(DE3) cells. Recombinant PPIB (34–216 aa), PLEC (175–400 aa), and EIF3A (8–848 aa) proteins with His-tags and StrepTag II corresponded to partial amino acid sequences. HNRNPR, FKBP4, HNRNPA2B1, HSPD1, PTBP1, PLEC, HNRNPA1, and FKBP1A were expressed as soluble fractions and purified using immobilized metal affinity chromatography (IMAC). RPL7A, EEF2K, LPCAT1, ENO1, KRT7, EEF1G, KRT19, EIF3A, TARDBP, KRT18, VIM, NY-ESO-1/CT6.1, and TCP1 were expressed as insoluble inclusion bodies and were solubilized using S-cationization techniques (Futami et al., 2015; Futami et al., 2017; Miyamoto et al., 2022; Murata et al., 2006). EMD, KRT8, and LMNA were refolded from their insoluble fractions without any chemical modifications. Purified recombinant autoantigens were identified as potential biomarker proteins based on positive western blot bands observed in diluted plasma from a patient with lung cancer.
4.7 Immunostaining of intracellular autoantigens
Subconfluent cells in a glass-based dish (Iwaki Glass, Shizuoka, Japan) were fixed with 4% paraformaldehyde phosphate buffer solution (Wako) and permeabilized with 0.1% Triton X-100 in PBS for 30 min. Intracellular autoantigens were reacted with 600-fold diluted human plasma in blocking buffer (1% BSA, 0.1% Tween20, in PBS) for 1 h at 25°C. Immunoreactive antigens and nuclei were stained with Alexa Fluor 488-conjugated AffiniPure Goat Anti-Human IgG (Jackson ImmunoResearch, West Grove, PA, USA, 1:400) and 4,6-diamidino-2-phenylindole (DAPI, Dojindo Laboratories, Kumamoto, Japan), respectively. Fluorescent images were acquired using a FV3000RS laser scanning microscope (Olympus, Tokyo, Japan) and an APX100 digital imaging system (EVIDENT, Tokyo, Japan).
4.8 Knockdown of endogenous specific autoantigens
Short hairpin RNAs (shRNAs) were constructed by ligating annealed oligo DNA into the pLKO.1 vector. For lentivirus preparation, 293T cells were transfected with 300 ng of pLKO.1-shRNA and packaging plasmids (300 ng of pMDLg/pRRE, 150 ng of pMD2g, and 150 ng of pRSVRev) using the FuGENE 6 transfection reagent (Promega, Madison, WI, USA) according to the manufacturer's instructions. Cells were infected using the filtered culture supernatant from 293T cells in the presence of 8 μg/mL polybrene. The target sequences of the shRNAs were 5′-GCTAACTACCAAGACACTATT-3′ for VIM, 5′-AGCTTCAGGTTATCGAAATAA-3′ for HNRNPA2B1, 5′-TCACTGTGTCCGGCAACATAC-3′ for NY-ESO-1, and 5′-CCTAAGGTTAAGTCGCCCTCG-3′ for the scrambled control.
4.9 Quantification of autoantibodies by MUSCAT-assay
Autoantibodies in patient plasma were diluted to 1/200 and quantified using water-soluble full-length antigen-immobilized Luminex beads. The detailed procedures have been described previously (Futami et al., 2015; Miyamoto et al., 2022).
AUTHOR CONTRIBUTIONS
Mirei Date, Ai Miyamoto, and Junichiro Futami conceived the project. Mirei Date, Ai Miyamoto, and Tomoko Honjo performed the autoantibody discovery experiments. Tsugumi Shiokawa and Hiroko Tada performed LC–MS/MS analysis. Nobuhiro Okada designed knockdown cells. Mirei Date and Junichiro Futami prepared the draft manuscript. All authors carried out revisions on the final manuscript version.
ACKNOWLEDGMENTS
This work is partially supported by JST START Grant Number JPMJST1918 (JF), JSPS KAKENHI Grant Number 22H01881 (JF), JST SPRING, Grant Number JPMJSP2126 (AM), and Science and Technology Promotion grants (2019–2023) in Okayama Prefecture, Japan.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.




