Assessing heterogeneity of electronic health-care databases: A case study of background incidence rates of venous thromboembolism
Abstract
Purpose
Heterogeneous results from multi-database studies have been observed, for example, in the context of generating background incidence rates (IRs) for adverse events of special interest for SARS-CoV-2 vaccines. In this study, we aimed to explore different between-database sources of heterogeneity influencing the estimated background IR of venous thromboembolism (VTE).
Methods
Through forest plots and random-effects models, we performed a qualitative and quantitative assessment of heterogeneity of VTE background IR derived from 11 databases from 6 European countries, using age and gender stratified background IR for the years 2017–2019 estimated in two studies. Sensitivity analyses were performed to assess the impact of selection criteria on the variability of the reported IR.
Results
A total of 54 257 284 subjects were included in this study. Age–gender pooled VTE IR varied from 5 to 421/100 000 person-years and IR increased with increasing age for both genders. Wide confidence intervals (CIs) demonstrated considerable within-data-source heterogeneity. Selecting databases with similar characteristics had only a minor impact on the variability as shown in forest plots and the magnitude of the I2 statistic, which remained large. Solely including databases with primary care and hospital data resulted in a noticeable decrease in heterogeneity.
Conclusions
Large variability in IR between data sources and within age group and gender strata warrants the need for stratification and limits the feasibility of a meaningful pooled estimate. A more detailed knowledge of the data characteristics, operationalisation of case definitions and cohort population might support an informed choice of the adequate databases to calculate reliable estimates.
Key Points
- Using a multi-database approach provides a more accurate picture of true IRs, as there may be large clinical differences underpinning the variability in the estimates across different databases.
- After mitigating unwanted heterogeneity through harmonization of database characteristics, there might still be some heterogeneity present, but this should be considered as a source of knowledge; our study confirmed prior knowledge that VTE backgrounds IRs were different dependents on the age and gender of the individual.
- The level of the heterogeneity in estimates depends on differences in database characteristics. In our study, databases collecting data from different parts of the health-care systems were the largest contributors to heterogeneity in estimates.
- When heterogeneity is present, a careful trade-off has to be made for the choice of IR, between stratified estimates or a pooled estimate, to support use in pharmacoepidemiological and regulatory evaluation.
- To attenuate heterogeneity, a pre-screening of database characteristics through a meta-dataset and adequate analytical tools at study design stage might be considered.
Plain Language Summary
Real-world data collected in everyday clinical practice can complement information used in regulatory decision-making and provide evidence to support the benefit-risk assessment of medicines. To improve the added value of real-world data for regulatory decision-making, regulators pool information from multiple databases to provide a more accurate picture of the outcome of interest. However, there is regularly variability, also called heterogeneity, in study outcomes when using data from different databases and this poses challenges for interpretation and communication. In this study, we examined incidence rates of venous thromboembolism, identified as a potential side effect for some COVID-19 vaccines, derived from multiple databases. We investigated how differences in database characteristics might cause variation between rate estimates and concluded that the largest contributor to heterogeneity was the use of data from different health-care settings. Understanding which database characteristics contribute to variability can allow to mitigate variation. This can be done by selecting databases with similar data characteristics, such as harmonised codes to refer to clinical outcomes and comparable selection criteria for participants and by using appropriate statistical methods to analyse the variability. Overall, our study provides an overview of the complexity of real-world evidence and can be used to better understand and analyse sources of variability.
1 INTRODUCTION
In the past two decades, the usage of large health-care databases has increased greatly.1 Regulatory agencies such as the European Medicines Agency (EMA) and the US Food and Drug Administration (FDA) have highlighted the value of real-world data (RWD) in medicines regulation.2, 3 In Europe, the initiation of the Data Analysis and Real World Interrogation Network (DARWIN EU),4 as well as the European Health Data Space are changing the landscape of real-world evidence (RWE) generation towards multi-database studies.
While there are already a number of advantages in using RWD in regulatory decision-making, those benefits can be improved by using more than one data source.5 Trivially, incorporating data from multiple data sources in an analysis will increase sample size. This can be crucial in situations with low event counts, such as for estimating the incidence rates (IRs) of a rare disease. While observational data are more generalizable to the real world than randomised controlled trials, the level of generalizability can be even further increased, by covering a broader and more representative population, thus possibly mitigating selection biases that are specific to single databases and by allowing for the quantification of true differences between populations.
Even though the benefits of using multiple data sources cannot be denied, data from those sources should not be pooled without a preliminary assessment of the suitability of pooling data, due to inherent differences in their characteristics. Simulations have shown increased risks of false-positive and false-negative safety signals when pooling data from multiple databases.6 This heterogeneity can have multiple forms, some of which are desirable for understanding true differences in outcomes event rates, while others make interpretation of results and decision-making regarding selection of suitable background rates for specific purposes such as observed-to-expected analyses for vaccines highly challenging.7
Sources of heterogeneity can be categorized into three types: measurement heterogeneity, information heterogeneity (both may be considered methodological heterogeneity), and true heterogeneity (also called clinical heterogeneity).8 While measurement (e.g., clinical classification systems) and information heterogeneity (e.g., granularity of clinical codes) can generally be considered undesirable, clinical heterogeneity has its value, for example, by improving external validity of results or understanding differences in prescription patterns or impact of risk minimisation measures (RMMs) between different geographical regions, health-care systems or behaviours. However, to understand heterogeneity, it is important to use appropriate tools to detect, report and account for it.
During the COVID-19 pandemic, RWD rapidly provided impactful evidence on safety and effectiveness of therapeutics and vaccines.9 This included the generation of background IR for adverse events of special interest (AESIs) for COVID-19 vaccines.10 Those background rates continue to be used in observed-to-expected analyses to estimate the expected number of cases in the general population prior introduction of COVID-19 vaccination, or during SARS-CoV2 circulation in non-vaccinated populations.
The list of AESIs included the concepts of deep vein thrombosis (DVT) and pulmonary embolism (PE). These two concepts make up the term venous thromboembolism (VTE).11 EMA pharmacovigilance activities identified VTE as a possible adverse event of Jcovden (former COVID-19 Vaccine Janssen)12 and listed VTE as an adverse event of Vaxzevria.13 Background rates were used to calculate the excess number of cases potentially linked with these vaccines. However, the reported background rates showed large differences between EU countries as reflected through national health-care records.14, 15
The objective of this study was to explore data characteristics that trigger heterogeneity in IR through both descriptive and statistical measures, using VTE as a case study. This investigation will further provide support in selecting adequate statistical methods for handling heterogeneity when pooling observations to derive meaningful pooled estimates to support regulatory decision-making.
2 METHODS
2.1 Data
To demonstrate an analytical workflow for handling heterogeneity between databases, we selected VTE, a safety concern listed for a class of COVID-19 vaccines, as a case study.
In order to assess potential adverse reactions related to approve COVID-19 vaccines in the EU, EMA-funded two studies through large research consortia: the ACCESS project with University Medical Center Utrecht and the EU PE&PV Research Network15, 16 and a study by ERASMUS University Medical Center17 to generate aggregated background IRs of AESIs, including VTE. Both consortia reported background IRs from multiple databases stratified by age group and gender, using the same eight age categories. IRs were estimated by dividing the number of incident cases by the total person-time at risk, with individuals entering the study cohort on their first visit after January 1, 2017, and being followed until the outcome, exit from the database or end of the study period. The study period covered 2017–2020. In the ACCESS protocol, the study population included all individuals who were observed in the databases for at least 1 day during the study period (January 1, 2017 to last date available) and who had at least 1 year of data availability before cohort entry, except for individuals <1 year of age with data available since birth. In the ERASMUS protocol, the study population was defined slightly different people observed on January 1 2017, January 1, 2018, or January 1, 2019 had to be observed continuously for at least 365 days with no event before this observation date. Ninety-five percent of CIs were calculated using an exact method described by Ulm.18
Case definitions for VTE were developed by the researchers independently. ACCESS utilized the CodeMapper tool19 to find harmonized definitions across coding systems. The full list of included concepts and details on its generation process is publicly available.20 Through using the OMOP common data model for its analyses, clinical codes in databases to which ERASMUS has access to were mapped to the SNOMED system, ensuring harmonized case definitions. The list of clinical codes included by ERASMUS can be accessed in the ATLAS application.21 Table A1 in the appendix shows included ICD10 codes for both ACCESS and ERASMUS. Only ICD10 codes are shown since both research organisations harmonized their definitions across coding systems. For both PE and DVT, the definition by ACCESS includes a broader range of concepts. For PE, the additional concepts are related to septic PE. The additional concepts for DVT mostly correspond to phlebitis, thrombophlebitis and DVT related to pregnancy. As described in a report by the FDA,22 there are no clear guidance on whether these concepts should be included or not.
A short overview of the databases is provided in Table 1. Further details, including demographic characteristics and total population, are provided in the corresponding published reports.15, 16 All databases are listed in the ENCePP research database, which also shows a list of relevant research publications they have been used in. For three databases (PHARMO, BIFAP, and SIDIAP), the IR had been estimated both on the total population and on the subset of subjects with linked primary care and hospital records (PC-H linkage). For the primary analysis, the total population estimates were used.
| Consortium | Region | Type of data source/health care system: Primary care (PC) or Hospital (H) data | Study population | Clinical Classification Coding system | |
|---|---|---|---|---|---|
| PHARMO | ACCESS | Netherlands | H | 9 184 832 | ICD10 |
| PHARMO | ACCESS | Netherlands | PC and H | 496 197 | ICPC (PC), ICD10 (H) |
| BIFAP | ACCESS | Spain | PC | 10 266 468 | SNOMED, ICD9 |
| BIFAP | ACCESS | Spain | PC and H | 4 423 843 | SNOMED, ICD9 (PC), ICD10 (H) |
| SIDIAP | ACCESS | Catalonia (Spain) | PC | 6 205 573 | ICD10 |
| SIDIAP | ACCESS | Catalonia (Spain) | PC and H | 1 758 239 | ICD10 |
| FISABIO | ACCESS | Valencia region (Spain) | PC and H | 5 886 560 | ICD9, ICD10 |
| PEDIANET | ACCESS | Italy | PC | 181 290 | ICD9 |
| ARS | ACCESS | Tuscany (Italy) | PC and H | 3 067 602 | ICD9 |
| CPRD GOLD | ACCESS | United Kingdom | PC | 4 688 710 | READ, SNOMED |
| CPRD GOLD | ERASMUS | United Kingdom | PC | 3 913 071 | READ |
| IQVIA DA Germany | ERASMUS | Germany | PC | 8 459 098 | ICD10 |
| IQVIA LPD France | ERASMUS | France | PC | 3 951 633 | ICD10 |
| IPCI | ERASMUS | Netherlands | PC | 1 299 288 | ICPC |
| IQVIA LPD Italy | ERASMUS | Italy | PC | 1 066 230 | ICD9 |
| SIDIAP | ERASMUS | Catalonia (Spain) | PC and H | 1 909 814 | ICD10 |
- Abbreviations: ARS, Agenzia Regionale di Sanita Toscana; BIFAP, Base de Datos para la Investigacion Farmacoepidemiologica en Atencian Primaria; CPRD, clinical practice research datalink; DA, disease analyzer; FISABIO, Fundación para el Fomento de la Investigación Sanitaria y Biomédica de la Comunitat Valenciana; IPCI, integrated primary care function; LPD, longitudinal patient data; SIDIAP, Sistema d'Informació per al Desenvolupament de la Investigació en Atenció Primària.
The data included the years 2017–2019. ACCESS reported IRs by year; hence, rates were pooled based on counts to match the data structure of ERASMUS, who reported only combined estimates for all years. Data from the Danish registries (DCE-AU) were reported only for the years 2010–2013 and thus were not included in the analysis. For the PHARMO database, only data for 2017 and 2019 was reported, due to an error in the imputation of a subset of data for 2018; BIFAP data with hospital linkage were only reported for 2017–2018.
2.2 Analysis
We used forest plots to visualize heterogeneity, displaying estimated IRs as squares with CIs for each database. The size of the squares is proportional to the precision of the estimate.
A random-effects meta-analysis, using the restricted maximum likelihood (REML) estimation method, was performed on the log scale of the IRs to calculate a summary estimate and to quantify the level of heterogeneity, thereby allowing for heterogeneity between databases, which is more realistic than assuming that the true value of the estimand is exactly the same for each database.
To quantify the absolute value of this heterogeneity, we reported estimates of τ2 measuring the ‘dispersion of true effect sizes between studies in terms of the scale of the effect size’23 and I2 measuring what proportion of variation in the observed effects is due to variation in true effects, that is, due to inherent differences between the investigated data sources rather than sampling error.24 Borenstein et al.18 stress that I2 represents a proportion rather than an absolute value. Therefore, we estimated the level of heterogeneity in comparison with statistical variability rather than heterogeneity itself. In addition, a prediction interval was calculated.25 Such an interval combines uncertainty due to sampling variation and due to heterogeneity to provide an approximate range of true values.
Finally, based on the available metadata characteristics of each of the included databases (see Table 1), several supplementary analyses were performed omitting or selecting a set of databases meeting selected criteria, aiming to reduce potential unwanted measurement and information heterogeneity, in order to assess more accurately true differences in subject-level data (i.e., true heterogeneity). These exploratory analyses allowed determining the contribution of each database characteristic to the heterogeneity in IR across databases.
- Restricting the analysis to a subpopulation; only those databases with linkage between primary care and hospital data can provide information on the influence of the health-care setting (i.e., type of data source). Including data sources with only primary care data might lead to underestimation in case of in-patient diagnosis. Data sources with only hospital data may underestimate the events in case of out-patient diagnosis.
- Restricting the analysis to only those databases using the same clinical classification system for diseases. In this study we included only databases that used the International Clinical Classification of Disease (ICD10)26 to diagnose VTE as it is the most widely used vocabulary among the available data sources.
- Restricting the analysis to databases with homogeneous case definition. Table A1 in the appendix specifies the ICD10 codes used to diagnose VTE in the two studies. We performed separate analyses by study, that is, ACCESS and ERASMUS, to explore differences in case definitions and population selection criteria.
- The analyses were performed using the R software27 package meta.28
3 RESULTS
A total of 60 080 169 subjects contributed to the 13 databases. See Tables A2 and A3 for an overview of the reported IRs stratified by age category and gender, by database and by study.
Two databases (CPRD GOLD and SIDIAP) were used in both studies by both consortia. Differences in defining the study cohort resulted in the cohort entry criteria not being identical. After removing the duplicated databases, a total of 54 257 284 subjects derived from 11 databases were included in the main analysis, representing collectively all age and gender subgroups from six countries.
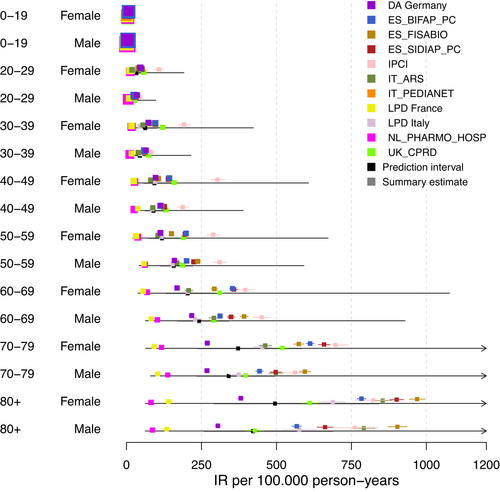
As the first step, we display the age–gender-database-specific IR estimates in a forest plot (Figure 1) using total population estimates. The forest plot showed a relatively large amount of heterogeneity between databases and within strata of age groups and gender. While for the 0–19 age group the IRs appeared to be in the same order of magnitude, with increasing age and increasing IRs the heterogeneity increased. In the 80+ age group, estimates ranged from <100 to >1 000 per 100 000 person-years. There did not seem to be a large difference in heterogeneity between gender categories. Two databases, LPD France and PHARMO, showed considerably lower estimates than the other databases in most age groups. The Dutch IPCI database, on the other hand, showed the highest estimates, especially for younger age groups and women. We generally observed consistent ranking of IR estimates across age groups, that is, across strata estimates are consistently high or low relative to the other databases.
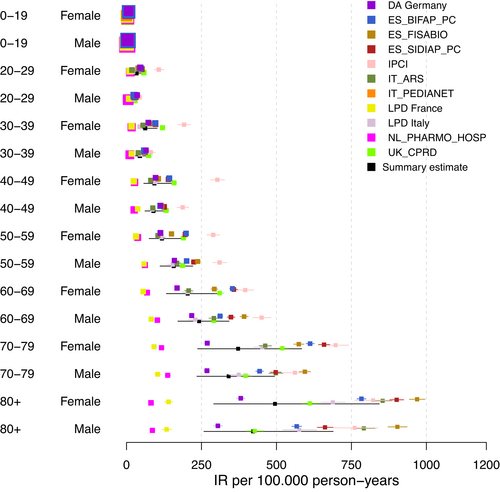
Next to the age–gender-database-specific IRs, age–gender IRs from meta-analyses were calculated. In our study, the meta-analysis estimated IR of VTE from 5 to 421 per 100 000 person-years depending on age–gender strata. The wide confidence interval of the summary (i.e., pooled) measure identified even within each stratum large patient-level differences. Table A4 in the appendix displays the age–gender IR estimates and CIs of the pooled measure for VTE from meta-analyses.
Figure A1 in the appendix shows the calculated prediction interval for the primary analysis. The prediction intervals for each age–gender group were notably high confirming the substantial population-level heterogeneity observed across data sources.
Table 2 shows the estimated I2 and τ2 values by age–gender stratum. The values for I2 indicated that a majority of the observed variability is due to differences between databases rather than random sampling error. Supporting the impression from the forest plot, we observed an increasing estimate of I2 with increasing age in both sexes. There did not seem to be an age-related trend in the estimates of τ2, but estimates for τ2 appear to be lower for males than for females.
| Age | Gender | I2 | τ2 |
|---|---|---|---|
| 0–19 | Female | 0.872 | 0.253 |
| 0–19 | Male | 0.921 | 0.376 |
| 20–29 | Female | 0.983 | 0.531 |
| 20–29 | Male | 0.954 | 0.381 |
| 30–39 | Female | 0.992 | 0.626 |
| 30–39 | Male | 0.981 | 0.423 |
| 40–49 | Female | 0.996 | 0.600 |
| 40–49 | Male | 0.991 | 0.367 |
| 50–59 | Female | 0.997 | 0.517 |
| 50–59 | Male | 0.995 | 0.299 |
| 60–69 | Female | 0.998 | 0.473 |
| 60–69 | Male | 0.996 | 0.309 |
| 70–79 | Female | 0.998 | 0.527 |
| 70–79 | Male | 0.997 | 0.358 |
| 80+ | Female | 0.998 | 0.735 |
| 80+ | Male | 0.997 | 0.625 |
3.1 Sensitivity analyses
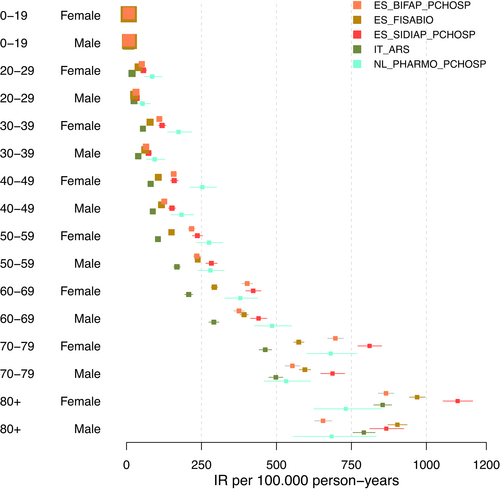
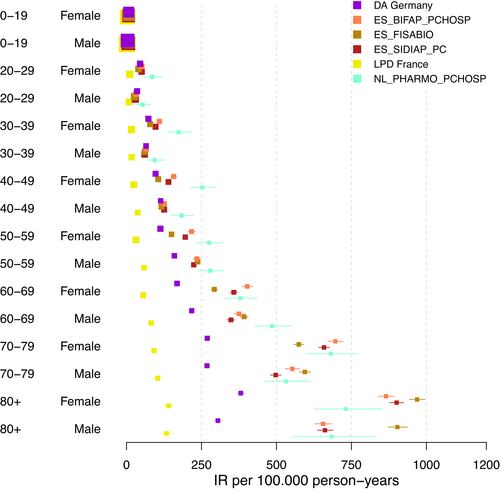
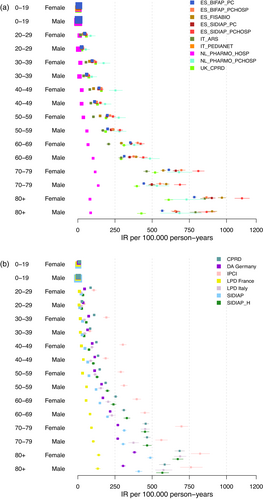
In the first sensitivity analysis, we restricted the databases to those with PC-H linkage (Figure 2). The forest plot demonstrates a relatively large decrease in heterogeneity when restricting the analysis to databases with PC-H linkage, across all age groups. It became apparent that this restriction of databases primarily leads to low estimates being excluded from the analysis. In Table A5 in the appendix, which lists I2 and τ2 estimates for all sensitivity analyses, we noticed lowered I2 estimates especially for younger age groups and considerably lowered τ2 values for all age groups. Figure 3 did not imply any reduction in heterogeneity when restricting the analysis to databases using ICD 10 codes to diagnose VTE. Both range and distribution of estimates were similar to the primary analysis. The same was true for estimates of I2 and τ2. Figure 4a, b showed forest plots of the analysis considering data from ACCESS and ERASMUS separately. Note that due to including the estimates from both the total population and the sub-population with PC-H linkage, there was some dependence between the estimates of BIFAP, SIDIAP, and PHARMO. For ACCESS, the hospital database PHARMO showed far lower estimates than all other databases included. This could be linked to an oversampling of the denominator. Apart from this, visually there seemed to be some reduction in heterogeneity. Data from ERASMUS and ACCESS showed a similar amount of heterogeneity; the forest plots indicate that estimates from ERASMUS are spread more equally, while the PHARMO hospital data differ strongly from the other estimates within ACCESS.
4 DISCUSSION
This study explored heterogeneity in background IRs of VTE reported from 11 data sources spanning six EU countries and derived from two observational studies, by focusing on the database as a source of heterogeneity. Through investigating data source characteristics potentially introducing differences in estimated IRs, our aim was to investigate the amount of unwanted (i.e., methodological) heterogeneity or uncertainty between data sources to provide more valid conclusions for safety surveillance activities. Data sources used in this study were mostly from primary care settings, partly with linkage to hospital data. The study used aggregated background IRs of VTE, considered a relevant AESI for a class of EU-approved COVID-19 vaccines.
Substantial heterogeneity in the background IRs was observed between all included data sources, in addition to observed within-data-source differences across age groups and genders. Age was the main contributor to the heterogeneity as shown in our study. Overall, it was observed that background rates increased with increasing age with no clear pattern in IR between males and females. The observation of increased IRs with increasing age is in line with another study on VTE,29 with the same study also suggesting a difference in IR between genders: IRs increase markedly with age for men and women; the overall age-adjusted IR is higher for men (130 per 100 000) than women (110 per 100 000). The observed heterogeneity in the different age–gender strata is a source of information that leads to a better understanding of the burden of VTE in the general population. However, as demonstrated through the summary estimate and CIs, we still found substantial heterogeneity between data sources within each stratum, suggesting that there still might be unobserved patient-level heterogeneity and therefore a single estimate for each stratum might be inaccurate.
In an attempt to understand the contribution of database characteristics to the reported heterogeneity, we performed several exploratory analyses. Our databases included data derived from both hospital and primary care settings. In all data sources, when estimating background rates, it is important to consider how the population denominator was derived. When linking data between the two settings, depending on the mechanism of linkage, there is a risk of only capturing those subjects that had a hospital visit recorded, which could lead to biased estimates. Restricting the databases that included a link with hospital (PC-H linkage) resulted in a moderate decrease in the reported variability. Alongside, we did not see a decrease in heterogeneity using only databases that used the ICD-10 vocabulary demonstrating that the type of vocabulary used for clinical classification of VTE could not be identified as a major source of heterogeneity. Differences in background rates remained between the two studies even if the time at risk in which the rates were collected and age–gender subgroup definitions and analytical methods were similar. Comparing more closely the methodology applied in the two studies, differences in case definitions were noted, with some clinical codes only included in one of the studies (Table A1 in the appendix). The inclusion and exclusion criteria for individuals also differed, leading to non-identical study populations even when within the same data source. Since we did not find any systematic differences in estimates between the two consortia, it is unlikely that differences in case definition or inclusion criteria had a large influence on observed heterogeneity.
When quantifying heterogeneity using the statistical measure I2, a considerable amount of heterogeneity (close to 100%) is reported. The large values of I2 are not surprising, as the large sample sizes in every database imply small variance estimates. In particular the I2 estimates in the 0–19 age group seem to be influenced by this fact: due to a larger sample size, the variance is lower than for the other age groups, leading to I2 estimates that appear too high in comparison with the other age groups when looking at the forest plot.
In this study, we have calculated pooled estimates for the primary analyses. However, when large heterogeneity is present, focusing on a pooled estimate is not advisable, given that the pooled estimate will derive largely from the particular choice of databases and the relative weights associated with each database.30 Following the classification in Deeks et al.8 it is not advised to combine the estimates if the value of I2 was estimated to be larger than 90%. In addition, when reporting the results after combining estimates, attention needs to be given to uncertainty quantification. In addition to the common risk of misinterpretation for CIs,31 CIs for random-effects meta-analyses are easily misinterpreted to quantify dispersion of study effects. However, CIs only represent the uncertainty in estimating the mean effect size, not taking into account variability due to different database characteristics. This means, that CIs are always smaller than the range of observed estimates.
A major strength of this study is the use of data aggregated from a large number of data sources independently provided by two research consortia using the same calculation method to estimate the IRs. This enabled the exploration of database-specific aspects related to heterogeneity. From 11 databases, 54 257 284 subjects contributed to the main analysis; with the databases spanning a large part of Europe, this can be considered a representative sample of the total population.
The above exploratory analyses show that even when certain database characteristics are harmonized, significant heterogeneity is still present. A limitation of our study is that only aggregated data was available which prevented us from investigating potential sources of heterogeneity attributable to patient characteristics. For instance, comorbidities may have an influence on IRs.32 A final limitation is that clinical validation of the diagnosis codes was not performed in these studies which might have led to different frequencies of misclassification or underreporting of VTE cases, which may affect estimates of heterogeneity.33
This study highlights the challenges regarding the varying levels of available information about database characteristics and the difficulty to identify sufficiently detailed information about the data sources. For example, some differences can only be explored through subject matter expertise about the corresponding health-care systems. Health-care systems might differ between regions, implying possible differences in the probability of recording certain events even in the same health-care setting. The process of clinical coding could also influence the quality of recording, with different levels of quality control or incentives for correct coding. With the large level of observed heterogeneity, an important recommendation is to use the same databases when comparing estimates at different time-points, for example, for pre- or post-exposure IRs of AESI.
The above considerations highlight the necessity for careful assessment of the suitability of databases to include in multi-database studies. In the two studies, a variety of databases was included because many different AESIs were considered simultaneously. For a study on a specific outcome, more specific restrictions on the databases should be placed a priori. In our study we have observed that the type of data source is one of the most important considerations. Based on subject matter knowledge or available validation studies, it should be evaluated in which type of setting the most accurate estimation is possible.
Our analysis also highlights the importance of unified case definitions and inclusion and exclusion cohort criteria to select adequate data sources in multi-database studies. This is evidenced in this study by the CPRD data source used by both study groups to address the same objective, but with a difference of 17% in the total number of individuals included in the study cohort, most likely related to differences in the operationalization of case definitions.
It is important to note that the methods used for detecting and addressing heterogeneity should be specified before starting any meta-analysis. When the forest plot show outliers among observed rates, it can be tempting to exclude the corresponding databases from the analysis without further investigating causes for outliers. This practice is, however, likely to introduce bias and should be avoided in most situations. Criteria for excluding certain databases should be specified prior to performing the analysis, but even then, it is advisable to also present results with the excluded databases, as a sensitivity analysis. In parallel, the choice of method for handling heterogeneity should also be prespecified, conditional of the outcome of the method for detecting heterogeneity. Also, it is preferable that the method for estimating the meta-analysis model and its statistical heterogeneity (e.g., REML), the methods for quantifying CIs (e.g., the Hartung-Knapp and Sidik-Jonkman modifications to the Wald method) and prediction intervals are prespecified.34
More specifically, exploring the level of heterogeneity using multiple databases must be considered if these rates are intended to be used to support safety signal detection activities and to avoid misleading recommendations. One of the current initiatives is the DIVERSE project with the aim to develop guidelines for the identification, collection and reporting of heterogeneity in multi-database studies.35 In addition, EMA's list of metadata for Real World Data catalogues,36 which will be the basis of a catalogue of RWD sources, will provide researchers with standardized, relevant information about databases to use for RWE studies. Another approach would be to develop a set of metrics to measure database heterogeneity or to develop phenotype libraries to identify important variables in different databases. For instance, Ostropolets et al.22 quantify factors influencing IRs through a set of sensitivity analysis using patient level data. Finally, the use of SNOMED CT (systematized nomenclature of medicine – clinical terms),37 a terminology that can cross-map to other classifications and code systems, may reduce the variability among estimates derived from different data sources. Nonetheless, the mapping of original coding systems to SNOMED may not reduce the heterogeneity as such, but may merely conceal possible heterogeneity introduced by different classification systems to operationalize the case definition of VTE. This is evidenced in this study by the ERASMUS data sources, where still large variability in the estimates is seen even when converted to SNOMED.
5 CONCLUSION
Our study revealed large variability in estimated age–gender-stratified background IRs of VTE between different databases, demonstrating the presence of one or several sources of heterogeneity. Restricting the databases to similar health-care settings contributed to less variability in the reported rates. Still, variability was present, triggered most likely by presence of analytical heterogeneity through differences in the case definitions and population cohorts as defined in the protocols used by the two study groups. The use of HARPER (harmonized protocol template to enhance reproducibility)38 to operationalize code definitions will improve the creation of unambiguous clinical codes in studies integrating data from multiple data sources.
Our study can be utilized to better understand the complexity of RWE and to illustrate the importance of a cautious selection of databases, based on their characteristics, so that the observed heterogeneity represents true differences, to ultimately improve the reliability of RWE. Our findings should be considered in context of similar analyses with other databases and in other settings.
AUTHOR CONTRIBUTIONS
All authors meet ICMJE criteria, they have all contributed to the development of the study, as well as writing and approval of the manuscript.
ACKNOWLEDGEMENTS
The authors thank Karin Hedenmalm and Luca Giraldi for their critical review of the manuscript.
DISCLAIMER
The views expressed in this article are the personal views of the authors and may not be understood or quoted as being made on behalf of or reflecting the position of the European Medicines Agency or one of its committees or working parties.
APPENDIX A
| Coding system | Code | Code name |
|---|---|---|
| ACCESSa | ||
| ICD10CM | I26 | pulmonary (acute) (artery)(vein) thromboembolism |
| ICD10CM | I26 | Pulmonary embolism |
| ICD10CM | I80 | Phlebitis and thrombophlebitis |
| ICD10CM | I81 | Portal vein thrombosis |
| ICD10CM | I82 | Other venous embolism and thrombosis |
| ICD10CM | O08.2 | Embolism following ectopic and molar pregnancy |
| ICD10CM | O22.3 | Deep phlebothrombosis in pregnancy |
| ICD10CM | O87.1 | Deep phlebothrombosis in the puerperium |
| Coding system | Code | Code name |
|---|---|---|
| ERASMUSb | ||
| ICD10 | I26 | Pulmonary embolism |
| ICD10CM | I26 | Pulmonary embolism |
| ICD10 | I26.0 | Pulmonary embolism with mention of acute cor pulmonale |
| ICD10CM | I26.0 | Pulmonary embolism with acute cor pulmonale |
| ICD10CM | I26.02 | Saddle embolus of pulmonary artery with acute cor pulmonale |
| ICD10CM | I26.09 | Other pulmonary embolism with acute cor pulmonale |
| ICD10 | I26.9 | Pulmonary embolism without mention of acute cor pulmonale |
| ICD10CM | I26.9 | Pulmonary embolism without acute cor pulmonale |
| ICD10CM | I26.92 | Saddle embolus of pulmonary artery without acute cor pulmonale |
| ICD10CM | I26.93 | Single subsegmental pulmonary embolism without acute cor pulmonale |
| ICD10CM | I26.94 | Multiple subsegmental pulmonary emboli without acute cor pulmonale |
| ICD10CM | I26.99 | Other pulmonary embolism without acute cor pulmonale |
| ICD10CM | I80.21 | Phlebitis and thrombophlebitis of iliac vein |
| ICD10CM | I80.219 | Phlebitis and thrombophlebitis of unspecified iliac vein |
| ICD10 | I82.2 | Embolism and thrombosis of vena cava |
| ICD10CM | I82.2 | Embolism and thrombosis of vena cava and other thoracic veins |
| ICD10CM | I82.21 | Embolism and thrombosis of superior vena cava |
| ICD10CM | I82.210 | Acute embolism and thrombosis of superior vena cava |
| ICD10CM | I82.211 | Chronic embolism and thrombosis of superior vena cava |
| ICD10CM | I82.22 | Embolism and thrombosis of inferior vena cava |
| ICD10CM | I82.220 | Acute embolism and thrombosis of inferior vena cava |
| ICD10CM | I82.221 | Chronic embolism and thrombosis of inferior vena cava |
| ICD10 | I82.3 | Embolism and thrombosis of renal vein |
| ICD10CM | I82.3 | Embolism and thrombosis of renal vein |
| ICD10CM | I82.4 | Acute embolism and thrombosis of deep veins of lower extremity |
| ICD10CM | I82.40 | Acute embolism and thrombosis of unspecified deep veins of lower extremity |
| ICD10CM | I82.401 | Acute embolism and thrombosis of unspecified deep veins of right lower extremity |
| ICD10CM | I82.402 | Acute embolism and thrombosis of unspecified deep veins of left lower extremity |
| ICD10CM | I82.403 | Acute embolism and thrombosis of unspecified deep veins of lower extremity, bilateral |
| ICD10CM | I82.409 | Acute embolism and thrombosis of unspecified deep veins of unspecified lower extremity |
| ICD10CM | I82.41 | Acute embolism and thrombosis of femoral vein |
| ICD10CM | I82.411 | Acute embolism and thrombosis of right femoral vein |
| ICD10CM | I82.412 | Acute embolism and thrombosis of left femoral vein |
| ICD10CM | I82.413 | Acute embolism and thrombosis of femoral vein, bilateral |
| ICD10CM | I82.419 | Acute embolism and thrombosis of unspecified femoral vein |
| ICD10CM | I82.42 | Acute embolism and thrombosis of iliac vein |
| ICD10CM | I82.421 | Acute embolism and thrombosis of right iliac vein |
| ICD10CM | I82.422 | Acute embolism and thrombosis of left iliac vein |
| ICD10CM | I82.423 | Acute embolism and thrombosis of iliac vein, bilateral |
| ICD10CM | I82.429 | Acute embolism and thrombosis of unspecified iliac vein |
| ICD10CM | I82.43 | Acute embolism and thrombosis of popliteal vein |
| ICD10CM | I82.431 | Acute embolism and thrombosis of right popliteal vein |
| ICD10CM | I82.432 | Acute embolism and thrombosis of left popliteal vein |
| ICD10CM | I82.433 | Acute embolism and thrombosis of popliteal vein, bilateral |
| ICD10CM | I82.439 | Acute embolism and thrombosis of unspecified popliteal vein |
| ICD10CM | I82.44 | Acute embolism and thrombosis of tibial vein |
| ICD10CM | I82.441 | Acute embolism and thrombosis of right tibial vein |
| ICD10CM | I82.442 | Acute embolism and thrombosis of left tibial vein |
| ICD10CM | I82.443 | Acute embolism and thrombosis of tibial vein, bilateral |
| ICD10CM | I82.449 | Acute embolism and thrombosis of unspecified tibial vein |
| ICD10CM | I82.49 | Acute embolism and thrombosis of other specified deep vein of lower extremity |
| ICD10CM | I82.491 | Acute embolism and thrombosis of other specified deep vein of right lower extremity |
| ICD10CM | I82.492 | Acute embolism and thrombosis of other specified deep vein of left lower extremity |
| ICD10CM | I82.493 | Acute embolism and thrombosis of other specified deep vein of lower extremity, bilateral |
| ICD10CM | I82.499 | Acute embolism and thrombosis of other specified deep vein of unspecified lower extremity |
| ICD10CM | I82.4Y | Acute embolism and thrombosis of unspecified deep veins of proximal lower extremity |
| ICD10CM | I82.4Y1 | Acute embolism and thrombosis of unspecified deep veins of right proximal lower extremity |
| ICD10CM | I82.4Y2 | Acute embolism and thrombosis of unspecified deep veins of left proximal lower extremity |
| ICD10CM | I82.4Y3 | Acute embolism and thrombosis of unspecified deep veins of proximal lower extremity, bilateral |
| ICD10CM | I82.4Y9 | Acute embolism and thrombosis of unspecified deep veins of unspecified proximal lower extremity |
| ICD10CM | I82.4Z | Acute embolism and thrombosis of unspecified deep veins of distal lower extremity |
| ICD10CM | I82.4Z1 | Acute embolism and thrombosis of unspecified deep veins of right distal lower extremity |
| ICD10CM | I82.4Z2 | Acute embolism and thrombosis of unspecified deep veins of left distal lower extremity |
| ICD10CM | I82.4Z3 | Acute embolism and thrombosis of unspecified deep veins of distal lower extremity, bilateral |
| ICD10CM | I82.4Z9 | Acute embolism and thrombosis of unspecified deep veins of unspecified distal lower extremity |
| ICD10CM | I82.5 | Chronic embolism and thrombosis of deep veins of lower extremity |
| ICD10CM | I82.50 | Chronic embolism and thrombosis of unspecified deep veins of lower extremity |
| ICD10CM | I82.501 | Chronic embolism and thrombosis of unspecified deep veins of right lower extremity |
| ICD10CM | I82.502 | Chronic embolism and thrombosis of unspecified deep veins of left lower extremity |
| ICD10CM | I82.503 | Chronic embolism and thrombosis of unspecified deep veins of lower extremity, bilateral |
| ICD10CM | I82.509 | Chronic embolism and thrombosis of unspecified deep veins of unspecified lower extremity |
| ICD10CM | I82.59 | Chronic embolism and thrombosis of other specified deep vein of lower extremity |
| ICD10CM | I82.591 | Chronic embolism and thrombosis of other specified deep vein of right lower extremity |
| ICD10CM | I82.592 | Chronic embolism and thrombosis of other specified deep vein of left lower extremity |
| ICD10CM | I82.593 | Chronic embolism and thrombosis of other specified deep vein of lower extremity, bilateral |
| ICD10CM | I82.599 | Chronic embolism and thrombosis of other specified deep vein of unspecified lower extremity |
| ICD10CM | I82.5Y | Chronic embolism and thrombosis of unspecified deep veins of proximal lower extremity |
| ICD10CM | I82.5Y1 | Chronic embolism and thrombosis of unspecified deep veins of right proximal lower extremity |
| ICD10CM | I82.5Y2 | Chronic embolism and thrombosis of unspecified deep veins of left proximal lower extremity |
| ICD10CM | I82.5Y3 | Chronic embolism and thrombosis of unspecified deep veins of proximal lower extremity, bilateral |
| ICD10CM | I82.5Y9 | Chronic embolism and thrombosis of unspecified deep veins of unspecified proximal lower extremity |
| ICD10CM | I82.62 | Acute embolism and thrombosis of deep veins of upper extremity |
| ICD10CM | I82.621 | Acute embolism and thrombosis of deep veins of right upper extremity |
| ICD10CM | I82.622 | Acute embolism and thrombosis of deep veins of left upper extremity |
| ICD10CM | I82.623 | Acute embolism and thrombosis of deep veins of upper extremity, bilateral |
| ICD10CM | I82.629 | Acute embolism and thrombosis of deep veins of unspecified upper extremity |
| ICD10CM | I82.81 | Embolism and thrombosis of superficial veins of lower extremities |
| ICD10CM | I82.811 | Embolism and thrombosis of superficial veins of right lower extremity |
| ICD10CM | I82.812 | Embolism and thrombosis of superficial veins of left lower extremity |
| ICD10CM | I82.813 | Embolism and thrombosis of superficial veins of lower extremities, bilateral |
| ICD10CM | I82.819 | Embolism and thrombosis of superficial veins of unspecified lower extremity |
| ICD10CM | I82.A | Embolism and thrombosis of axillary vein |
| ICD10CM | I82.A1 | Acute embolism and thrombosis of axillary vein |
| ICD10CM | I82.A11 | Acute embolism and thrombosis of right axillary vein |
| ICD10CM | I82.A12 | Acute embolism and thrombosis of left axillary vein |
| ICD10CM | I82.A13 | Acute embolism and thrombosis of axillary vein, bilateral |
| ICD10CM | I82.A19 | Acute embolism and thrombosis of unspecified axillary vein |
| ICD10CM | I82.B | Embolism and thrombosis of subclavian vein |
| ICD10CM | I82.B1 | Acute embolism and thrombosis of subclavian vein |
| ICD10CM | I82.B11 | Acute embolism and thrombosis of right subclavian vein |
| ICD10CM | I82.B12 | Acute embolism and thrombosis of left subclavian vein |
| ICD10CM | I82.B13 | Acute embolism and thrombosis of subclavian vein, bilateral |
| ICD10CM | I82.B19 | Acute embolism and thrombosis of unspecified subclavian vein |
| ICD10CM | I82.C | Embolism and thrombosis of internal jugular vein |
| ICD10CM | I82.C1 | Acute embolism and thrombosis of internal jugular vein |
| ICD10CM | I82.C11 | Acute embolism and thrombosis of right internal jugular vein |
| ICD10CM | I82.C12 | Acute embolism and thrombosis of left internal jugular vein |
| ICD10CM | I82.C13 | Acute embolism and thrombosis of internal jugular vein, bilateral |
| ICD10CM | I82.C19 | Acute embolism and thrombosis of unspecified internal jugular vein |
- a ACCESS included all subcodes, whereas ERASMUS only used the listed codes and not any unlisted codes.
- b ICD-10 codes not included by ERASMUS as compared to ACCESS are: I26.01, I26.90, I80 and subcodes except I80.21 and I80.219, I81 and subcodes, I82, I82.0, I82.1, I82.29, I82.290, I82.291, I82.51-I82.56 and I82.5Z and subcodes, I82.6-I82.7 and I82.9 and subcodes except I82.62 and subcodes, I82.B2 and I82.C2 and subcodes O08.02, O22.3, O87.1.
| SIDIAP_H | IR_upr | 8.401 | 8.558 | 42.831 | 34.043 | 65.147 | 50.089 | 83.982 | 114.790 | 142.046 | 214.288 | 257.495 | 351.061 | 479.374 | 495.524 | 708.428 | 611.455 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IR_lwr | 3.683 | 3.935 | 27.519 | 20.090 | 49.371 | 35.527 | 66.827 | 94.446 | 117.867 | 184.456 | 222.594 | 309.225 | 425.596 | 437.671 | 637.169 | 528.101 | |
| IR | 5.691 | 5.921 | 34.551 | 26.391 | 56.852 | 42.344 | 75.039 | 104.247 | 129.535 | 198.953 | 239.569 | 329.646 | 451.886 | 465.925 | 672.090 | 568.635 | |
| SIDIAP | IR_upr | 3.965 | 2.158 | 26.031 | 16.999 | 35.844 | 33.165 | 52.101 | 71.404 | 80.333 | 129.702 | 156.578 | 209.455 | 325.792 | 319.963 | 506.255 | 432.463 |
| IR_lwr | 2.114 | 0.874 | 19.069 | 11.348 | 28.958 | 26.360 | 44.267 | 62.204 | 69.831 | 115.971 | 140.480 | 189.719 | 298.792 | 290.318 | 469.686 | 387.497 | |
| IR | 2.933 | 1.412 | 22.349 | 13.962 | 32.264 | 29.616 | 48.065 | 66.685 | 74.945 | 122.692 | 148.365 | 199.404 | 312.073 | 304.870 | 487.713 | 409.517 | |
| LPD ITALY | IR_upr | 18.533 | 20.361 | 29.521 | 35.718 | 56.030 | 60.506 | 108.185 | 102.275 | 129.371 | 173.693 | 222.919 | 254.114 | 482.714 | 409.460 | 728.965 | 633.780 |
| IR_lwr | 3.125 | 3.973 | 13.952 | 15.924 | 35.379 | 34.264 | 83.512 | 72.635 | 103.875 | 136.809 | 186.667 | 206.169 | 424.357 | 341.139 | 648.693 | 522.077 | |
| IR | 8.515 | 9.882 | 20.679 | 24.377 | 44.830 | 46.019 | 95.254 | 86.515 | 116.101 | 154.431 | 204.192 | 229.207 | 452.832 | 374.134 | 687.952 | 575.907 | |
| LPD FRANCE | IR_upr | 3.078 | 2.431 | 14.368 | 12.986 | 20.481 | 22.188 | 28.467 | 43.741 | 36.220 | 65.696 | 61.570 | 90.441 | 100.744 | 114.658 | 153.878 | 150.461 |
| IR_lwr | 0.803 | 0.474 | 7.661 | 5.259 | 13.299 | 12.565 | 20.533 | 31.514 | 27.612 | 51.922 | 49.968 | 74.395 | 83.309 | 93.896 | 127.632 | 118.459 | |
| IR | 1.674 | 1.180 | 10.629 | 8.496 | 16.604 | 16.875 | 24.259 | 37.256 | 31.699 | 58.507 | 55.543 | 82.125 | 91.717 | 103.890 | 140.297 | 133.747 | |
| IPCI | IR_upr | 18.781 | 9.74 | 124.661 | 53.590 | 213.464 | 95.729 | 326.663 | 207.130 | 310.978 | 333.740 | 424.226 | 481.229 | 740.676 | 603.106 | 886.734 | 837.853 |
| IR_lwr | 10.165 | 3.853 | 92.163 | 33.065 | 171.910 | 66.879 | 280.467 | 168.438 | 268.142 | 288.068 | 369.025 | 421.215 | 655.350 | 522.530 | 761.507 | 689.366 | |
| IR | 13.990 | 6.308 | 107.500 | 42.416 | 191.848 | 80.346 | 302.907 | 187.038 | 288.966 | 310.275 | 395.905 | 450.474 | 697.035 | 561.737 | 822.337 | 760.906 | |
| DA Germany | IR_upr | 8.974 | 4.870 | 50.190 | 41.029 | 78.340 | 71.979 | 102.490 | 121.669 | 118.153 | 166.851 | 175.389 | 226.173 | 278.467 | 278.472 | 394.823 | 319.480 |
| IR_lwr | 5.575 | 2.439 | 40.803 | 30.197 | 67.861 | 58.776 | 91.478 | 106.356 | 108.009 | 152.551 | 161.889 | 208.809 | 260.228 | 258.221 | 367.633 | 290.473 | |
| IR | 7.126 | 3.502 | 45.315 | 35.305 | 72.960 | 65.128 | 96.867 | 113.819 | 112.995 | 159.581 | 168.538 | 217.361 | 269.231 | 268.203 | 381.046 | 304.717 | |
| CPRD | IR_upr | 13.048 | 4.148 | 94.268 | 32.978 | 118.780 | 90.853 | 133.897 | 145.978 | 156.620 | 197.799 | 268.255 | 324.970 | 477.566 | 488.495 | 717.938 | 622.249 |
| IR_lwr | 8.636 | 1.849 | 77.761 | 23.299 | 101.457 | 74.995 | 116.096 | 126.803 | 138.019 | 176.771 | 240.892 | 294.459 | 435.655 | 443.471 | 652.590 | 547.731 | |
| IR | 10.674 | 2.831 | 85.718 | 27.827 | 109.863 | 82.640 | 124.759 | 136.138 | 147.099 | 187.064 | 254.297 | 309.433 | 456.250 | 465.575 | 684.679 | 584.100 | |
| gender | Female | Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | Male | |
| age_gr | 0–19 | 0–19 | 20–29 | 20–29 | 30–39 | 30–39 | 40–49 | 40–49 | 50–59 | 50–59 | 60–69 | 60–69 | 70–79 | 70–79 | 80+ | 80+ |
| IT_PEDIANET | IR_upr | 2.781 | 3.872 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IR_lwr | 0 | 0.018 | |||||||||||||||
| IR | 0 | 0.695 | |||||||||||||||
| UK_CPRD | IR_upr | 4.880 | 3.650 | 64.120 | 30.740 | 128.380 | 80.630 | 168.040 | 140.790 | 200.070 | 196.850 | 325.070 | 305.510 | 541.160 | 416.750 | 639.560 | 457.340 |
| IR_lwr | 2.580 | 1.740 | 51.520 | 22.360 | 111.950 | 67.700 | 149.330 | 123.930 | 180.140 | 177.260 | 296.480 | 277.500 | 499.000 | 377.750 | 583.400 | 399.390 | |
| IR | 3.600 | 2.570 | 57.570 | 26.300 | 119.960 | 73.950 | 158.480 | 132.160 | 189.910 | 186.860 | 310.530 | 291.250 | 519.760 | 396.890 | 610.990 | 427.630 | |
| SIDIAP | IR | 2.930 | 1.410 | 22.350 | 13.960 | 32.260 | 29.620 | 48.060 | 66.690 | 74.940 | 122.690 | 148.370 | 199.400 | 312.070 | 304.870 | 487.710 | 409.520 |
| NL_PHARMO_PCHOSP | IR_upr | 33.180 | 26.760 | 117.900 | 79.800 | 216.990 | 127.630 | 300.050 | 224.150 | 321.150 | 325.390 | 436.910 | 549.590 | 768.650 | 613.540 | 850.010 | 832.610 |
| IR_lwr | 10.810 | 7.470 | 60.230 | 33.710 | 136.590 | 66.960 | 211.410 | 148.350 | 234.340 | 238.820 | 327.790 | 426.980 | 601.030 | 459.250 | 625.020 | 555.830 | |
| IR | 19.780 | 14.960 | 85.540 | 53.180 | 173.360 | 93.730 | 252.860 | 183.360 | 275.210 | 279.620 | 379.440 | 485.410 | 681.000 | 532.240 | 731.090 | 683.890 | |
| NL_PHARMO_HOSP | IR_upr | 5.490 | 3.320 | 14.800 | 7.100 | 19.500 | 13.270 | 29.410 | 28.410 | 40.710 | 65.520 | 73.750 | 109.220 | 124.250 | 146.060 | 87.750 | 94.100 |
| IR_lwr | 3.240 | 1.660 | 10.540 | 4.110 | 14.850 | 9.190 | 23.840 | 22.650 | 34.100 | 56.820 | 63.780 | 96.600 | 109.440 | 128.960 | 75.850 | 78.940 | |
| IR | 4.260 | 2.390 | 12.530 | 5.460 | 17.050 | 11.090 | 26.520 | 25.410 | 37.300 | 61.050 | 68.630 | 102.760 | 116.670 | 137.310 | 81.640 | 86.280 | |
| IT_ARS | IR_upr | 7.960 | 9.680 | 24.190 | 31.090 | 63.250 | 46.430 | 88.570 | 96.200 | 113.730 | 179.620 | 221.040 | 308.280 | 483.490 | 521.680 | 884.070 | 828.570 |
| IR_lwr | 3.600 | 4.950 | 13.700 | 19.510 | 47.120 | 32.580 | 73.010 | 79.640 | 96.410 | 157.230 | 194.180 | 274.950 | 442.240 | 475.000 | 824.970 | 755.530 | |
| IR | 5.470 | 7.030 | 18.400 | 24.800 | 54.740 | 39.050 | 80.510 | 87.630 | 104.800 | 168.140 | 207.290 | 291.260 | 462.520 | 497.930 | 854.140 | 791.420 | |
| ES_SIDIAP_PCHOSP | IR_upr | 13.200 | 12.910 | 66.990 | 42.210 | 131.210 | 83.290 | 172.120 | 163.870 | 253.610 | 301.970 | 448.040 | 467.950 | 850.560 | 727.520 | 1154.280 | 924.290 |
| IR_lwr | 6.920 | 6.880 | 46.800 | 26.820 | 106.880 | 64.330 | 146.590 | 139.830 | 219.720 | 265.000 | 397.970 | 413.990 | 771.920 | 648.100 | 1055.760 | 811.170 | |
| IR | 9.690 | 9.550 | 56.220 | 33.870 | 118.580 | 73.350 | 158.970 | 151.490 | 236.210 | 283.030 | 422.450 | 440.350 | 810.520 | 686.950 | 1104.190 | 866.350 | |
| ES_SIDIAP_PC | IR_upr | 8.370 | 7.380 | 55.390 | 33.200 | 102.730 | 65.050 | 146.040 | 131.130 | 203.940 | 232.820 | 370.020 | 360.970 | 677.690 | 515.690 | 924.290 | 688.350 |
| IR_lwr | 5.730 | 4.970 | 45.710 | 25.880 | 91.470 | 56.240 | 133.670 | 119.820 | 187.910 | 215.680 | 345.810 | 335.930 | 640.150 | 479.820 | 878.070 | 636.240 | |
| IR | 6.960 | 6.090 | 50.380 | 29.370 | 96.980 | 60.520 | 139.750 | 125.380 | 195.800 | 224.130 | 357.760 | 348.280 | 658.720 | 497.510 | 900.960 | 661.910 | |
| ES_FISABIO | IR_upr | 8.930 | 11.360 | 44.490 | 30.200 | 84.110 | 66.510 | 111.720 | 122.540 | 157.240 | 246.670 | 303.970 | 405.680 | 591.720 | 614.000 | 995.190 | 935.500 |
| IR_lwr | 6.110 | 8.240 | 35.450 | 22.890 | 73.280 | 56.930 | 100.300 | 110.840 | 142.870 | 228.450 | 282.010 | 378.810 | 557.590 | 576.050 | 943.420 | 871.940 | |
| IR | 7.420 | 9.700 | 39.780 | 26.350 | 78.560 | 61.580 | 105.900 | 116.580 | 149.930 | 237.430 | 292.840 | 392.070 | 574.470 | 594.800 | 969.050 | 903.300 | |
| ES_BIFAP_PCHOSP | IR_upr | 10.140 | 9.450 | 58.450 | 37.620 | 118.620 | 72.350 | 167.140 | 134.890 | 228.930 | 246.740 | 420.460 | 392.630 | 722.730 | 578.150 | 891.520 | 684.130 |
| IR_lwr | 5.900 | 5.460 | 44.020 | 26.210 | 100.760 | 58.420 | 147.770 | 117.620 | 205.620 | 222.610 | 384.980 | 358.170 | 670.840 | 528.900 | 839.780 | 627.510 | |
| IR | 7.810 | 7.250 | 50.850 | 31.530 | 109.420 | 65.110 | 157.230 | 126.030 | 217.040 | 234.440 | 402.430 | 375.100 | 696.430 | 553.110 | 865.360 | 655.360 | |
| ES_BIFAP_PC | IR_upr | 7.980 | 5.980 | 49.790 | 28.510 | 100.760 | 63.040 | 147.680 | 117.380 | 204.810 | 206.660 | 362.580 | 320.880 | 625.820 | 456.320 | 798.300 | 583.870 |
| IR_lwr | 5.890 | 4.230 | 42.430 | 23.010 | 91.690 | 55.840 | 137.640 | 108.470 | 192.540 | 194.280 | 344.240 | 303.160 | 598.780 | 431.280 | 768.780 | 551.730 | |
| IR | 6.880 | 5.050 | 46.000 | 25.650 | 96.150 | 59.360 | 142.590 | 112.860 | 198.600 | 200.400 | 353.320 | 311.920 | 612.190 | 443.670 | 783.430 | 567.630 | |
| gender | Female | Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | Male | |
| age_gr | 0–19 | 0–19 | 20–29 | 20–29 | 30–39 | 30–39 | 40–49 | 40–49 | 50–59 | 50–59 | 60–69 | 60–69 | 70–79 | 70–79 | 80+ | 80+ |
| Age | Gender | Estimate | Lower | Upper |
|---|---|---|---|---|
| 0–19 | Female | 5.697 | 4.095 | 7.924 |
| 0–19 | Male | 4.322 | 2.911 | 6.418 |
| 20–29 | Female | 32.773 | 20.772 | 51.707 |
| 20–29 | Male | 21.825 | 14.778 | 32.231 |
| 30–39 | Female | 62.316 | 38.087 | 101.957 |
| 30–39 | Male | 44.485 | 29.621 | 66.807 |
| 40–49 | Female | 93.155 | 57.581 | 150.707 |
| 40–49 | Male | 89.761 | 61.579 | 130.841 |
| 50–59 | Female | 118.074 | 75.574 | 184.476 |
| 50–59 | Male | 157.258 | 111.909 | 220.984 |
| 60–69 | Female | 203.979 | 133.110 | 312.582 |
| 60–69 | Male | 242.021 | 171.372 | 341.798 |
| 70–79 | Female | 372.142 | 237.184 | 583.891 |
| 70–79 | Male | 340.595 | 234.922 | 493.800 |
| 80+ | Female | 495.147 | 290.933 | 842.703 |
| 80+ | Male | 421.791 | 258.186 | 689.069 |
| PC-H linkage | ICD-10 code | ACCESS | ERASMUS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Age | Gender | I2 | τ2 | I2 | τ2 | I2 | τ2 | I2 | τ2 |
| 0–19 | Female | 0.771 | 0.145 | 0.894 | 0.506 | 0.851 | 0.177 | 0.944 | 0.521 |
| 0–19 | Male | 0.539 | 0.019 | 0.928 | 0.588 | 0.927 | 0.309 | 0.896 | 0.479 |
| 20–29 | Female | 0.942 | 0.293 | 0.961 | 0.328 | 0.975 | 0.345 | 0.988 | 0.573 |
| 20–29 | Male | 0.731 | 0.042 | 0.936 | 0.245 | 0.950 | 0.351 | 0.966 | 0.290 |
| 30–39 | Female | 0.971 | 0.178 | 0.985 | 0.481 | 0.990 | 0.443 | 0.994 | 0.570 |
| 30–39 | Male | 0.904 | 0.084 | 0.954 | 0.208 | 0.980 | 0.383 | 0.983 | 0.250 |
| 40–49 | Female | 0.985 | 0.184 | 0.991 | 0.485 | 0.993 | 0.406 | 0.996 | 0.554 |
| 40–49 | Male | 0.958 | 0.070 | 0.976 | 0.204 | 0.990 | 0.322 | 0.990 | 0.235 |
| 50–59 | Female | 0.988 | 0.149 | 0.994 | 0.463 | 0.995 | 0.366 | 0.995 | 0.392 |
| 50–59 | Male | 0.970 | 0.042 | 0.991 | 0.218 | 0.994 | 0.213 | 0.992 | 0.199 |
| 60–69 | Female | 0.989 | 0.086 | 0.997 | 0.445 | 0.996 | 0.321 | 0.995 | 0.305 |
| 60–69 | Male | 0.968 | 0.035 | 0.995 | 0.286 | 0.995 | 0.205 | 0.994 | 0.215 |
| 70–79 | Female | 0.988 | 0.045 | 0.998 | 0.468 | 0.997 | 0.335 | 0.996 | 0.304 |
| 70–79 | Male | 0.951 | 0.014 | 0.997 | 0.320 | 0.999 | 0.226 | 0.994 | 0.232 |
| 80+ | Female | 0.967 | 0.019 | 0.998 | 0.368 | 0.998 | 0.631 | 0.994 | 0.228 |
| 80+ | Male | 0.970 | 0.018 | 0.997 | 0.343 | 0.997 | 0.521 | 0.991 | 0.222 |




