Novel markers and networks related to restored skeletal muscle transcriptome after bariatric surgery
Abstract
Objective
The aim of this study was to discover novel markers underlying the improvement of skeletal muscle metabolism after bariatric surgery.
Methods
Skeletal muscle transcriptome data of lean people and people with obesity, before and 1 year after bariatric surgery, were subjected to weighted gene co-expression network analysis (WGCNA) and least absolute shrinkage and selection operator (LASSO) regression. Results of LASSO were confirmed in a replication cohort.
Results
The expression levels of 440 genes differing between individuals with and without obesity were no longer different 1 year after surgery, indicating restoration. WGCNA clustered 116 genes with normalized expression in one major module, particularly correlating to weight loss and decreased plasma free fatty acids (FFA), 44 of which showed an obesity-related phenotype upon deletion in mice. Among the genes of the major module, 105 represented prominent markers for reduced FFA concentration, including 55 marker genes for decreased BMI in both the discovery and replication cohorts.
Conclusions
Previously unknown gene networks and marker genes underlined the important role of FFA in restoring muscle gene expression after bariatric surgery and further suggest novel therapeutic targets for obesity.
Study Importance
What is already known?
- Bariatric surgery is an effective method for treatment of obesity, resulting in improved insulin sensitivity accompanied by changes in skeletal muscle gene expression.
What does this study add?
- Here, we discovered novel multidimensional links between molecular and metabolic changes induced by bariatric surgery. The study identified novel networks and prominent markers for changes in BMI and plasma free fatty acids.
How might these results change the direction of research or the focus of clinical practice?
- The prominent markers of BMI and plasma free fatty acid improvement can be used for the investigation of future therapeutic targets in the treatment of obesity.
INTRODUCTION
At present, bariatric or metabolic surgery remains the most effective and durable treatment for obesity and can even delay the progression from prediabetes to type 2 diabetes (T2D) [(1, 2)]. Although many metabolic and molecular changes occurring after bariatric surgery have been described, the investigation of markers associated with improvements in body weight loss and metabolism has been limited.
Skeletal muscle is mainly responsible for insulin-stimulated glucose disposal and thereby determines whole-body insulin sensitivity in humans [(3-5)]. Recently, we closely monitored the changes of systemic, as well as muscle, metabolism and gene expression over the first 52 weeks after bariatric surgery. This time-course analysis demonstrated that the progressive improvement of whole-body insulin sensitivity is paralleled by changes in the expression of genes involved in lipid metabolism, mitochondrial functionality, and insulin signaling [(6)].
Until now, few studies have performed genome-wide transcriptome analyses of skeletal muscle after bariatric surgery [(7-10)]. However, they did not include detailed phenotypical characterizations such as clamp studies or in-depth comprehensive bioinformatic analyses of genome-wide transcriptome. For instance, Barres et al. described that weight loss, within 6 months after bariatric surgery, results in restoration of expression of genes related to metabolic and mitochondrial pathways to levels observed in lean humans [(8)]. In another study, only one candidate, integrin β-3 (ITGB3), was shown to reach expression levels observed in lean humans at 3 months after bariatric surgery [(7)].
Even though many groups have investigated bariatric surgery outcomes, a comprehensive connection between changes in metabolic traits and the variation in gene expression networks is still missing. To this end, we used a bioinformatic strategy including weighted gene co-expression network analysis (WGCNA) and least absolute shrinkage and selection operator (LASSO) regression and validated the results of LASSO with RNA sequencing analysis, which we performed with samples of a replication cohort. We particularly focused on transcripts and epigenetic changes that are corrected to the levels of lean control individuals, evaluated their gene networks and hub genes, and, finally, identified biomarkers of weight loss and decreased free fatty acids (FFA) concentration after surgery. Of these, candidates with altered DNA methylation that are also, according to the literature, detectable in blood cells may possibly serve as easily available biomarkers for both successful body weight loss and improvement of fatty acid metabolism.
METHODS
Study population
Discovery cohort
The present analysis comprised a subgroup of people from a previous report [(6)] of the Bariatric surgery–Improvement of basic requirements for targeted therapy (BARIA-DDZ) cohort study, which monitors individuals before and over the course of 5 years after bariatric surgery with comprehensive phenotyping [(11)]. Detailed information on the participants and experimental protocols have been reported elsewhere [(6)]. In brief, 16 participants were studied before (people with obesity at week 0 [OB w0]) and 52 weeks after (people with obesity at week 52 [OB w52]) surgery (sleeve gastrectomy or gastric bypass), and 7 healthy, lean, age-matched people were examined once to serve as the control group (CON). All participants of the discovery and replication cohort underwent anthropometric measurements (including body mass index [BMI]), fasting blood sampling, a 3-h hyperinsulinemic-euglycemic clamp, and a biopsy of vastus lateralis muscle at each visit [(12)]. Adiponectin, FFA, fasting glucose levels, and glycated hemoglobin (HbA1c) were measured in CON, OB w0, and OB w52.
Replication cohort
To replicate the discovery cohort results, we used data of another group of 13 female participants with obesity before and 52 weeks after surgery. The phenotypic traits were identical to those used for the discovery cohort (Figure S1 and Tables S1 and S2). Participants from both cohorts are all of German-European ancestry.
All study participants received information about all procedures and risks before providing their written consent to the protocol (clinical trial registration: ClinicalTrials.gov NCT01477957). Approval was provided by ethics committees at the Faculty of Medicine of Heinrich Heine University and the Medical Association North Rhine (AEKNO), Düsseldorf, Germany.
Mitochondrial function
Muscle biopsy samples were taken from the vastus lateralis muscle under local anesthesia [(13)]. Ex vivo analysis of mitochondrial respiratory capacity was performed on permeabilized muscle fibers in a two-chamber oxygraph (OROBOROS Instruments, Innsbruck, Austria), as described previously [(14)]. In permeabilized muscle fibers, β oxidation-linked respiratory rates were analyzed by subsequent addition of malate, octanoyl-carnitine, ADP, glutamate, and succinate. Then, cytochrome c was added to test the integrity of the outer mitochondrial membrane, followed by incremental titration steps of 1.0 μL carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) (0.1 mmol/L) until maximal uncoupled respiration was achieved.
Gene expression
For the discovery cohort, we used our previously generated data, which are publicly available via Gene Expression Omnibus (GEO)-Number GSE135066. Data of the replication cohort were not published previously. Thirty milligrams of skeletal muscle biopsies of 13 participants were used for RNA isolation, as described [(6)]. The RNA sequencing analysis was carried out by BGI Genomics Laboratory in Hong Kong, China, via nanoball sequencing technology (Table S1).
DNA methylation
For the discovery cohort, we reanalyzed our previously generated DNA methylation data in skeletal muscle biopsies, publicly available via GEO-Number GSE135066 (detailed in Table S1). To verify whether genes affected by DNA methylation changes in skeletal muscle after bariatric surgery were also epigenetically regulated in blood cells, we extracted data from a publicly available database, Epigenome-Wide Association Studies (EWAS) Atlas [(15, 16)], and compared it with our candidates. Key terms such as “BMI” and “BMI change” were applied to the EWAS search. The hypergeometric test was applied to the resulting overlap to exclude risks of random intersection.
Gene ontology
Gene ontology analyses were performed as described in (17)]. The category selected was biological processes, and the parameters were set to the following: at least five genes per gene ontology term, minimum fold enrichment of two, and p < 0.05.
WGCNA
WGCNA is a systems biology method for depicting correlation patterns among differentially expressed genes and identifies clusters (modules) of highly correlated genes [(11)]. We applied WGCNA with the following parameters: power = 8 (Figure S2); minModuleSize = 30; networktype = unsigned; and cutHeight = 0.25. The WGCNA pipeline was applied to gene expression data, as described in (18)]. Functions of the WGCNA package [(19)] were used in R version 3.6.1 (Vienna, Austria). To link modules to phenotypes, the module eigengenes were calculated and correlated (Spearman correlation) with the phenotype data and were then corrected for multiple testing by false discovery rate.
Receptor-ligand interactions
Potential receptor-ligand interactions among the gene products of the M1 module were collected from CellPhoneDB [(20)].
Secretome analysis
Potentially secreted gene products were identified through publicly available Vertebrate Secretome Database (VerSeDa) and the Human Protein Atlas (HPA) (https://www.proteinatlas.org/) data.
Mouse knockout databases
Genes identified in module M1 were screened for obesity-related phenotypes obtained in knockout mice that were analyzed by the International Mouse Phenotyping Consortium (IMPC) and Mouse Genome Informatics (MGI) [(21, 22)]. Data were collected in March 2021.
LASSO analysis
LASSO regression analysis is a machine-learning approach that allows identification of prognostic markers by performing variable selection and regularization to enhance the prediction accuracy [(12, 13)]. LASSO regression was performed using transcriptome data and phenotypes of individuals with and without obesity before and 52 weeks after surgery. The data set was randomly split into a test (50%) and train-data set (50%). R version 4.1.3 was used with R-package “glmnet” version 4.1. λ was estimated using a five-fold cross validation. One thousand models were generated, and a model was supposed to be successful if 50% of the phenotypes were predicted within a range of 10%. Genes of successful models were collected and then combined to identify those candidates that have a high potential as markers for a chosen phenotype. This approach lowers the probability of artificial effects and identifies marker genes without overfitting.
Statistical analysis
Programming and calculation of statistics for transcriptome were performed with R version 4.0.3 and 4.1.2. Raw p values were calculated by two-tailed Welch's t test for transcriptome.
R-packages “ggplots,” “ComplexHeatmap,” “circlize,” and “plotly” were used to generate heat maps, a Sanky diagram, and cyclized plots, respectively.
RESULTS
Study design and cohorts
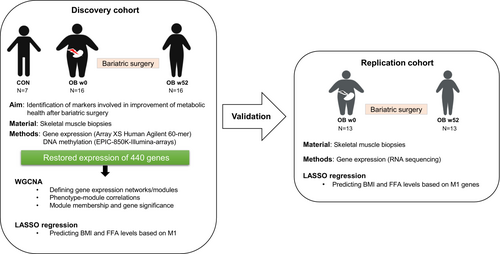
Figure 1 and Figure S1 show the study design, including the different steps and cohorts used for the identification of gene networks and markers of skeletal muscle, which may participate in the improvement of metabolic health after bariatric surgery. For the first part of the study, data of a discovery cohort (7 people without and 16 with obesity) were analyzed. In a second part, data of a replication cohort (13 people with obesity) were used for the verification of the findings obtained by LASSO regression. Table 1 summarizes the anthropometric data of the study participants of discovery cohort, and Table S2 shows the corresponding data of the replication cohort.
| CON | OB w0 | OB w52 | |
|---|---|---|---|
| N (male) | 7 (2) | 16 (5) | 16 (5) |
| Age (y) | 38.4 ± 9.3 | 38.7 ± 9.4 | 38.7 ± 9.4 |
| BMI (kg/m2) | 24.9 ± 2.7 | 49.5 ± 7.0* | 32.0 ± 5.0*,** |
| ΔBMI (%) | - | - | 35.0 ± 7.8 |
| M-value | 7.6 ± 2.1 | 2.7 ± 1.5* | 6.3 ± 2.0** |
| Fasting glucose (mg/dL) | 75.3 ± 5.0 | 94.2 ± 22.9* | 76.5 ± 9.1** |
| Fasting FFA (μmol/L) | 518.4 ± 189.3 | 710.2 ± 160.2 | 554.6 ± 158.5** |
| Total adiponectin (ng/mL) | 5899.5 ± 1095.0 | 5085.9 ± 1845.0 | 8719.6 ± 2537.7*,** |
| HMW-adiponectin (ng/mL) | 3549.2 ± 1244.5 | 2233.2 ± 1323.1 | 4937.1 ± 2078.2** |
| Sleeve gastrectomy/gastric bypass | - | - | 8/8 |
- Note: Data given as mean ± SD.
- Abbreviations: CON, control lean persons; FFA, free fatty acids; HMW, high molecular weight; OB w0, people with obesity at week 0; OB w52, people with obesity at week 52.
- * p < 0.05 vs. CON using unpaired t test with Welch's correction.
- ** p < 0.05 vs. OB w0 using paired t test.
After bariatric surgery, the participants of the discovery cohort showed a decrease in BMI, blood glucose, and fasting plasma FFA by 35%, 19%, and 22%, respectively, whereas the M-value (clamp-derived whole-body insulin sensitivity) increased by 57%. Both baseline data and changes after surgery did not differ between those undergoing gastric bypass or sleeve gastrectomy (data not shown).
The majority of differences in muscle gene expression between lean people and people with obesity disappears after bariatric surgery
In order to evaluate to which extent muscle transcriptome reverted to levels of those observed in lean people 1 year after bariatric surgery, array-based transcriptome data were assessed in muscle biopsies of participants with obesity (OB) and lean control people (CON). Before surgery, 580 genes were differentially expressed (differentially expressed genes [DEG]) in skeletal muscle between OB w0 and CON (Table S3). After 52 weeks, expression of 440 genes (76%) was no longer different between OB w52 and CON, whereas 140 genes remained significantly altered Figure 2A,B, Figure S1B, and Table S4. No significant differences were observed between male and female expression patterns in the whole transcriptome or in the normalized genes (Figure S3). Similar results were obtained for the type of surgery (Figure 2B and Figure S4A). Gene ontology analysis on the 440 normalized genes revealed a significant enrichment for 207 genes involved in inflammatory response, protein kinase B activation, fatty acid metabolism, regulation of autophagy, phosphoinositide 3-kinases (PI3K) signaling, and others (Table S5 and Figure S4B).
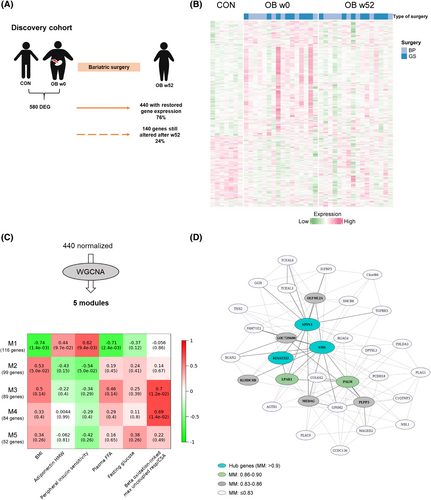
We have already previously shown that DNA methylation was linked to transcriptional changes in response bariatric surgery [(6)]. Therefore, we examined DNA methylation profiles of the 440 normalized genes. As shown in Figure S5A, 322 genes (1785 CpG sites with adjusted p < 0.05) were affected by DNA methylation in skeletal muscle after bariatric surgery. The heat maps depict strong differences in the DNA methylation patterns in CON and OB w0 and show a relatively similar pattern between CON and OB w52 (Figure S5B). To verify whether the same genes were affected by epigenetic alterations in an easily accessible tissue, DNA methylation data from blood cells were collected from the EWAS Atlas [(15, 16)] and then compared with the 322 differentially methylated DEG. Out of these, 54 also showed epigenetic alterations in blood cells (hypergeometric test, p < 6 × 10−7; Figure S5A), and their methylation patterns were significantly associated with BMI and/or BMI changes in previous studies (Table S6) [(15)]. Taken together, a majority of the genes with expression that reached the levels of people without obesity after bariatric surgery also show adjustments in their DNA methylation.
Specific gene networks associate with surgical improvement of insulin sensitivity
Considering that obesity is caused by a dysregulated interplay of multiple genes, we tested which of the genes with normalized expression are co-expressed and thereby build specific networks by applying WGCNA. The 440 normalized genes were grouped into five modules based on similarities in their expression profiles (Figure 2C). Module M1 was the largest, with 116 genes, of which spindlin1 (SPIN1), spermine synthase (SMS), and uncharacterized protein KIAA1522 were identified as hub genes because they had the strongest connections in this module (Figure 2C,D). The top 10 genes exhibiting a connectivity above 0.83 (Table S7), including the three hub genes, are marked in Figure 2D. Correlations of each module with the clinical traits (Table 1) are shown in Figure 2C. Module M1 displayed the strongest correlations to the phenotypic features of decreased BMI, improved whole-body insulin sensitivity, and reduced FFA concentrations (r = −0.74, 0.62, and −0.71, respectively) and was also linked to increased adiponectin levels (r = 0.44) and reduced fasting blood glucose (r = −0.37; Figure 2C). Both M3 and M4 showed specific correlation to β oxidation-linked maximum uncoupled respiration (r = 0.7 for M3 and r = 0.69 for M4; Figure 2C). To get at least indirect evidence for the function of several genes within the modules, we implemented phenotype data from respective knockout mice collected from the IMPC database [(21)]. In M1, out of 116 candidates, 59 were listed with a corresponding knockout mouse, and, among these, 44 showed an obesity-related phenotype (Figure 3, Figure S6A, and Table S8). As shown in Figure 3, the deletion of 18 genes led to significant changes in body mass, 30 affected glucose homeostasis, 2 affected plasma insulin concentration, and 22 affected the concentration of plasma lipids (e.g., triglycerides and cholesterol). Among these were also the hub genes SMS and SPIN1. Mice lacking Sms were leaner than wild-type littermates and exhibited altered circulating glucose concentrations. SPIN1 knockout mice showed preweaning lethality, and heterozygous mice exhibited elevated fasting blood glucose levels (Figure 3). Using the MGI database, we identified an additional 17 genes that have been linked to relevant phenotypes, among others with susceptibility to weight loss (e.g., acyl-CoA synthetase short chain family member 1 [Acss1]), circulating glucose levels (e.g., insulin-like growth factor binding protein 3 [Igfbp3]), energy expenditure (e.g., free fatty acid receptor 4 [Ffar4]), and skeletal muscle morphology (Table S8).
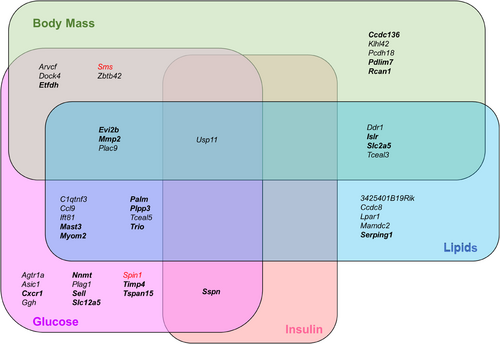
Even though M3 and M4 genes were enriched in inflammation and phosphatidylinositol signaling pathways (Figure S6B), many genes were linked to obesity phenotypes according to knockout mouse data. Out of 89 genes in M3 and 84 genes in M4, 22 and 24 genes associated with changes in body mass (e.g., Rho GTPase activating protein 25 [Arhgap25] and angiogenin, ribonuclease A family, member 6 [Ang6]), glucose homeostasis (e.g., GTP dependent ribosome recycling factor mitochondrial 2 [Gfm2] and C-X-C motif chemokine ligand 9 [Cxcl9]), insulin (e.g., leukocyte specific transcript 1 [Lst1] and coactosin like F-actin binding protein 1 [Cotl1]), and plasma lipids (e.g., sirtuin 2 [Sirt2] and RAP1 GTPase activating protein 2 [Rap1gap2]) in whole-body mouse knockout models, respectively (Figure S6C,D).
These analyses indicate that the normalized genes cluster in specific modules, which are associated with favorable postsurgical phenotypes.
Characterization of genes of module M1
Genes of the M1 module appear to include the most promising genes with a restored expression and strongly correlate to the improvement of muscle metabolism and weight loss after bariatric surgery. The majority of M1 genes were not described either in obesity genome-wide association (0/116 genes) or gene expression quantification loci (16/116 genes) studies (Table S9). Thus, the current analysis put a specific focus on novel genes, unknown in previous genetic studies, as a putative contributor in the improvement of metabolic health after bariatric surgery. We therefore conducted additional analyses with the 116 M1 genes.
The network depicted in Figure 2D shows several genes encoding for receptors (e.g., transforming growth factor β receptor 3 [TGFBR3] and angiotensin II type 1 receptor [AGTR1]) and secreted proteins (IGFBP3) with high connectivity in M1. Module membership (MM) coefficient was high for the aforementioned genes listed, ranging between 0.78 and 0.77 (Table S7). Screening all M1 genes for products potentially mediating ligand-receptor interactions with CellPhoneDB, we identified two genes encoding for ligands and six for receptors, suggesting 20 putative receptor-ligand interactions (Figure 4A).
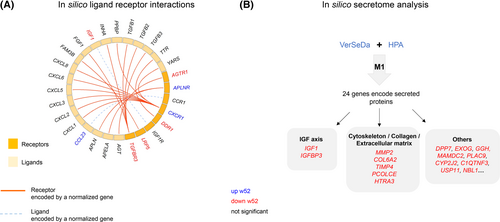
To identify genes encoding for secreted proteins that may improve muscle and whole-body metabolism on the level of organ cross talk, the 116 M1 genes were analyzed via the VerSeDa [(23)] and HPA databases. This approach identified 24 candidates, partially overlapping with results of CellPhoneDB (Figure 4B). Those 24 genes were linked to different cellular and molecular functions: two candidates act in the insulin-like growth factor (IGF) axis, and five are related to cytoskeleton and extracellular matrix composition (Table S10).
Those putative interactions determined additional signaling pathways that were not detected by gene ontology analysis. For instance, TGFBR3 expression was normalized at week 52, and CellPhoneDB predicted fibroblast growth factor 1 (FGF1) as its potential ligand (Figure 4A). This can be linked to the Smad signaling pathway, which has been related to insulin resistance-induced muscle atrophy [(24)].
Therefore, a large number of genes belonging to the M1 module are already known to play a role in obesity-related metabolic alterations, and some of them may be involved in an improved receptor-mediated signaling and/or the cross talk with other tissues. However, M1 also contains genes that have not been linked to physiological changes induced by bariatric surgery.
Plasma FFA and BMI are the main features correlated with M1
To estimate the relevance of each gene belonging to the M1 module in relation to tested clinical traits, the network properties MM, which defines the degree of connectivity of each gene with other candidates in the network, and “gene significance” (GS) were calculated [(25)]. The correlation between MM and GS reflects whether highly connected genes in a module are indeed strongly correlated to a selected phenotype. Among the five clinical traits shown in Figure 2C, the strongest correlation was found for FFA (r = 0.71, p = 8 × 10−18; Figure 5A, right panel). Weaker correlations were detected for BMI (r = 0.29, p = 2 × 10−3; Figure 5A, left panel) and M-value (r = 0.19, p = 0.049), and no significant correlations were found for glucose and adiponectin levels (Figure S7). Thus, the expression of M1-genes may be considered as prominent marker for changes in FFA and BMI. Therefore, both FFA and BMI were selected as main features for LASSO regression analysis.
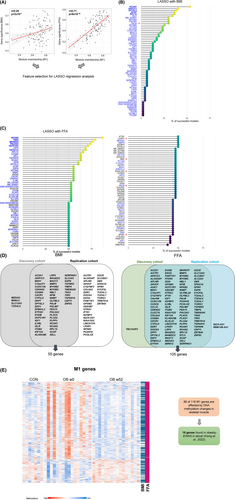
LASSO regression revealed novel marker genes linked to postsurgical changes of FFA and BMI
To identify putative markers responsible for improvement of specific obesity-related traits after bariatric surgery, LASSO regression was applied to M1 genes. First, 1000 random prediction models were built for BMI, as well as for FFA, using M1 gene expression data. A generated model is designated as successful if the calculated phenotype values were less than 10% different from the measured ones in at least 50% of the samples. Finally, genes within the M1 module were ranked based on how many models identified them to be predictive for changes in BMI (Figure 5B) and FFA (Figure 5C). As expected from GS and MM correlations (Figure 5A), LASSO regression resulted in a higher number of markers for FFA (108 genes) than BMI (60 genes). Of note, 60 M1 genes were indicators for both BMI and FFA concentrations (Figure 5B,C). For example, regulator of calcineurin 2 (RCAN2) appeared in 87% of the successful models indicating FFA concentrations and in 57% models for BMI changes. Hub genes and those candidates with the highest connectivity according to WGCNA (lysophosphatidic acid receptor 1 [LPAR1], SMS, paralemmin [PALM], SPIN1, and KIAA1522) are marked by stars in Figure 5C. Little is known about most of the strongly connected genes, as well as the highly predictive genes, identified by LASSO (RCAN2, tensin 2 [TNS2], MAGE family member D2 [MAGED2], pleckstrin homology like domain family A member 3 [PHLDA3], and host cell factor C1 regulator 1 [HCFC1R1]). Only RCAN2, TNS2, and SMS have been previously described in relation to obesity or skeletal muscle function [(26-29)]. By reanalyzing publicly available muscle transcriptome data, several candidates were found to be linked to obesity [(30, 31)]. Among the markers for FFA, 20 were significantly altered in skeletal muscle of monozygotic twin pairs discordant for BMI (Table S11).
Validation of LASSO regression analysis in an independent obesity cohort
To exclude certain biases and limitations of the current analysis (e.g., power, overfitting), we used expression data from an independent group of participants who underwent bariatric surgery (Figure 1, Figure S1, and Table S1) in order to validate the findings of the LASSO regression. For this, the 116 genes of the M1 module were analyzed in skeletal muscle expression data of a group of 13 people with obesity before and after bariatric surgery. Of 116 M1 genes identified by array-based transcriptome analysis in the discovery cohort, 109 genes could be mapped to results obtained by RNA sequencing analysis. These genes were utilized for LASSO regression, with BMI and FFA changes after bariatric surgery resulting in the identification of 107 markers for FFA levels and 88 for BMI (Table S12). As shown in the Venn diagrams, genes linked to BMI (55 genes) and FFA (105) identified in the discovery cohort strongly overlapped with those detected in the replication cohort (Figure 5D). Thus, results obtained in the discovery cohort were confirmed in the replication cohort. Taken together, 55 and 105 genes, which exhibit restored expression in skeletal muscle 1 year after bariatric surgery, are prominent markers for the decrease in BMI and circulating FFA, respectively. Because expression of genes in skeletal muscle are not easy to be used as prediction markers, we tested which of the DEG with changes in DNA methylation (Figure 5E) show alterations in blood cells [(15)] and may thereby be used as epigenetic markers. Among the 116 M1 genes, 19 candidates, including the hub (e.g., PALM and KIAA1522) and marker genes (e.g., RCAN1, SREBF chaperone [SCAP], and PDZ and LIM domain 7 [PDLIM7]), exhibit differential DNA methylations in blood cells of people with and without obesity, suggesting that they are useful biomarkers for weight loss and improvement of insulin sensitivity (Table 2).
| Gene symbol | Protein name | UniProt function |
|---|---|---|
| BDH1 | 3-hydroxybutyrate dehydrogenase 1 | - |
| COL6A2 | Collagen type VI α 2 chain | Cell-binding protein |
| CXCR1 | C-X-C motif chemokine receptor 1 | Receptor to interleukin 8 |
| EEF2 | Eukaryotic translation elongation factor 2 | Catalyzes ribosomal translocation during translation elongation |
| EXOG | Exo/endonuclease G | Endo/exonuclease |
| GGH | γ-glutamyl hydrolase | Hydrolyzes the polyglutamate side chains of pteroylpolyglutamates |
| IGFBP3 | Insulin-like growth factor binding protein 3 | Binds IGF and modulates its receptor interactions |
| KIAA1522 | KIAA1522 | - |
| KIF13A | Kinesin family member 13A | Regulates intracellular transport |
| LRP5 | LDL receptor related protein 5 | Acts as a coreceptor to transduce signal by Wnt proteins |
| NBL1 | NBL1, DAN family BMP antagonist | Possible tumor suppressor |
| NFIB | Nuclear factor I B | Transcriptional activator of GFAP |
| PALM | Paralemmin | Involved in plasma membrane dynamics |
| PDLIM7 | PDZ and LIM domain 7 | May function as a scaffold for protein assembly |
| PLAG1 | PLAG1 zinc finger | Transcriptional activator |
| RCAN1 | Regulator of calcineurin 1 | Inhibits calcineurin-dependent transcriptional responses |
| SCAP | SREBF chaperone | Escort protein required for cholesterol and lipid homeostasis |
| SERPING1 | Serpin family G member 1 | May play a role in regulation of physiological pathways |
| TRIO | Trio Rho guanine nucleotide exchange factor | Guanine nucleotide exchange factor for RHOA and RAC1 GTPases |
- Abbreviation: EWAS, epigenome-wide association studies.
DISCUSSION
In comparison with our recent publication [(6)], the present study focuses on transcripts and epigenetic changes, which are reaching the levels of those observed in lean people, and applied comprehensive bioinformatic analyses on the corresponding transcriptome and methylome data. As a result, we identified networks and novel hub genes and now provide a list of marker genes predicting successful weight loss and/or reduction of circulating FFA concentrations. Out data demonstrate that the majority (76%) of transcriptional alterations detected in skeletal muscle of people with obesity are restored to levels of those observed in lean people within 1 year after bariatric surgery. Machine-learning analysis revealed multidimensional links between phenotypic alterations and variations in gene expression that have not been discovered in previous studies. Our bioinformatic approaches performed with the 440 restored genes indicate that they clustered into five modules based on their transcriptional similarities, of which, one module (M1) with 116 genes showed the strongest correlation to BMI, whole-body insulin sensitivity, and plasma FFA. Of these 116 genes, 88 exhibited a restored DNA methylation pattern. M1 genes include candidates linked to putative receptor-ligand interactions and myokines, and 61 M1 genes are involved in the regulation of body weight, glucose and lipid metabolism, and insulin action according to results obtained by corresponding knockout mice. Finally, LASSO regression detected 55 and 105 M1 genes to be prominent markers for BMI and plasma FFA, respectively, suggesting that the decrease in circulating FFA concentration after surgery-mediated weight loss may play a particular role in restoring gene expression in skeletal muscle.
Although gene ontology and pathway enrichment analyses showed their robustness over the last decade, they have limitations. For instance, enrichment analyses usually cover only a subset of genes. In fact, gene ontology enrichment of the actual study assigned only 207 of the 440 genes to biological pathways. Among the 440 genes, only 4 genes showed an overlap with results obtained by Barres et al. [(8)], who investigated expression changes in skeletal muscle 6 months after bariatric surgery. This indicates that a longer period of surgery-induced weight loss is required for normalization of altered expression pattern.
Here, we adapted a unique bioinformatic strategy to clarify how transcriptional changes participate in the amelioration of the overall muscle metabolism. To the best of our knowledge, none of the previous studies had used WGCNA and LASSO to analyze the transcriptional alterations after bariatric surgery.
The WGCNA, which combines numerous genes with similar expression levels to networks, demonstrated that not only single genes but gene networks are restored to levels of people without obesity after surgery. In particular, expression of genes belonging to the M1 module strongly correlated with BMI, whole-body insulin sensitivity, and plasma FFA. Except for SMS, the M1 hub genes KIAA1522 and SPIN1 have not been associated with obesity or insulin sensitivity thus far but may be interesting candidates for future investigation. SMS was shown to have beneficial metabolic effects in high-fat diet-induced obese mice, such as by lowering fat mass and plasma lipids, attenuating hepatic steatosis, and decreasing inflammatory cytokine and chemokine expression in adipose tissue [(32)]. Another M1 hub gene was SPIN1, encoding a histone methylation reader, which epigenetically controls multiple tumorigenesis-associated signaling pathways, including the Wnt and PI3K/protein kinase B (AKT) pathways [(33)]. A mouse model with specific Spin1 deletion in myoblast precursors showed an aberrant myogenesis, abnormal glycogen metabolism, and neuromuscular junction defects [(34)]. Thus, the increase of SPIN1 expression in the muscle after surgery might have beneficial effects on the global gene expression and muscle regeneration in people with obesity.
Module M1 connects its hub genes to candidates that encode for receptors and/or ligands, such as TGFBR3 and IGFBP3. FGF1, a ligand of TGFBR3, mediates multiple beneficial effects in obesity and diabetes, such as glucose lowering [(35)], by enhancing cell surface localization of glucose transporter type 4 (GLUT4) [(36)], indicating that the elevated TGFBR3 expression participates in improved glucose homeostasis.
LASSO allowed the identification of putative markers playing a particular role for decreasing BMI and circulating FFA 1 year after bariatric surgery. Among the top genes detected, RCAN2, TNS2, and PHLDA3 belong to important signaling pathways. RCAN2 is a calcineurin inhibitor that plays a role in the development of obesity. Its deletion in mice suppressed diet-induced obesity [(26)]. TNS2 encodes for a tyrosine-protein phosphatase, which affects the stability of insulin receptor substrate 1 (IRS1) in skeletal muscle [(27)]. In HEK293 cells, TNS2 was described as a negative regulator of AKT/PKB signaling, thereby affecting cell proliferation and migration [(28, 29)]. Therefore, we speculate that the normalization of TNS2 expression can be linked to the improvement of AKT/PKB and/or IRS1 signaling after surgically induced weight loss. PHLDA3 encodes a phosphatidylinositol binding protein known to interfere with AKT translocation to the plasma membrane [(37)]. Taken together, our results, in combination with earlier findings described in literature, indicate that RCAN2, TNS2, and PHLDA3 are major candidates mediating the improvement of muscle metabolism after bariatric surgery.
To validate our findings, we tested expression data of an independent group of participants with obesity from the German Diabetes Center (DDZ) as a replication cohort. Unfortunately, we could not include other publicly available transcriptome data described in literature because they lack data on plasma FFA concentrations. Despite successful validation, a limitation of our study is that it was conducted in two cohorts from a population of German ancestry. Therefore, currently, our findings cannot be transferred to people with different genetic backgrounds. Additionally, mechanistic insights of the identified markers were not addressed in the current study.
CONCLUSION
Surgical weight loss restores the expression of most affected genes in skeletal muscle of people with obesity to levels of lean control individuals. In addition to common metabolic pathways, this study identified novel networks and prominent marker genes as predictors for changes in BMI and circulating FFA. Furthermore, the study indicates that the identified markers are particularly affected by decreasing circulating FFA.
AUTHOR CONTRIBUTIONS
Meriem Ouni, Michael Roden, and Annette Schürmann performed study conception and design. Meriem Ouni and Leona Kovac performed data acquisition for this manuscript. Sofiya Gancheva and Sabine Kahl provided and analyzed the clinical data. Meriem Ouni, Leona Kovac, Markus Jähnert, Erika Zuljan, Annette Schürmann, and Pascal Gottmann carried out bioinformatic analysis. Meriem Ouni and Annette Schürmann wrote the paper. Michael Roden is the principal investigator of the Bariatric surgery–Improvement of basic requirements for targeted therapy (BARIA-DDZ) study. Annette Schürmann has primary responsibility for the final content of this manuscript. All authors read, critically revised, and approved the final version of the manuscript.
ACKNOWLEDGMENTS
The authors thank Franziska Gabler, Eva Arlt, and Fariba Zivehe for their excellent technical assistance. Open Access funding enabled and organized by Projekt DEAL.
FUNDING INFORMATION
The work was supported by the German Ministry of Education and Research (BMBF: DZD grant 82DZD03D03), German Research Foundation (DFG: 491394008), and the Brandenburg State. This study was supported in part by the Ministry of Culture and Science of the State of North Rhine-Westfalia (MKW NRW), the German Federal Ministry of Health (BMG), and by a grant of the German Ministry of Education and Research to the German Center for Diabetes Research (DZD e.V., DZD Grant 2016). Michael Roden is further supported by grants from the German Science Foundation (CRC/SFB1116/2 B12; RTG/GRK 2576 vivid, Project 3) and the Schmutzler Stiftung. Sabine Kahl has received research grants from the German Center for Diabetes Research and the German Diabetes Society (DDG).
CONFLICT OF INTEREST STATEMENT
The authors declared no conflict of interest.





