Genome-Wide Scan of Fifth Finger Clinodactyly
Funding: Recruitment and phenotyping for the COHRA2 and OFC2 projects were funded by grants from the National Institute for Dental and Craniofacial Research (NIDCR): R01-DE014899 (M.L.M.) and R01-DE016148 (M.L.M., S.M.W.), respectively. Genotyping services were funded by grants from the National Institutes of Health (NIH): X01-HG009878 (J.R.S.) and X01-HG011437 (J.R.S., M.L.M.), respectively. The GABC project was funded in part by a grant from the National Heart, Lung, and Blood Institute (NHLBI): R37-HL039693. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ABSTRACT
Background
Fifth finger clinodactyly describes the conspicuous curvature of the fifth digit toward the other digits of the hand. Phenotypic expression can range from mild and almost imperceptible to severe, where function is impacted, and clinical intervention may be required. Although classically considered an autosomal dominant trait based on early family studies, no genes have been mapped for the trait. Further, there is epidemiological evidence that mild (typical-range) fifth finger clinodactyly may have a different etiology than more severe forms.
Methods
In this retrospective cross-sectional study, we carried out genome-wide association mapping of common genetic variants for clinodactyly in three cohorts separately and combined results via meta-analysis, treating the trait as either a continuous quantitative variable (nmeta = 631) or a binary outcome (nmeta = 1647).
Results
The vast majority of participants in these cohorts exhibited mild forms of clinodactyly. Both the individual cohort results and meta-analyses revealed no genome-wide significant loci. We identified several possible suggestive signals (p < 1 × 10−6), but these showed no evidence of replication.
Conclusion
While our results cannot definitively exclude the contribution of common variants to fifth finger clinodactyly due to the small sample size, they do suggest that the mild form of the trait is unlikely to be related to a major gene effect operating in a simple Mendelian manner.
1 Introduction
Clinodactyly describes a curvature of the finger(s) in the coronal plane, most often involving a deviation of the fifth finger toward the radial side of the forelimb (Biesecker et al. 2009). In rare instances, the curvature is severe enough to require therapeutic intervention, but most cases are mild and unproblematic (Flatt 2005). In a recent report (Lee et al. 2022), we showed that some degree of radial fifth finger curvature was universally present in 1295 adults; the mean curvature was just over 3.5° and only 0.5% of individuals exceeded the 10° threshold for clinically defined clinodactyly. Because there are few, if any, functional consequences, mild clinodactyly has received little attention despite its ubiquity. Most studies focus on severe forms of the condition. Severe fifth finger clinodactyly is believed to involve an abnormality of the growth plate between the middle and distal phalanges (Rayan and Upton 2014; Moeller 2019), is frequently reported in certain genetic syndromes (e.g., Down Syndrome, OMIM: 190685; Flatt 2005), and, based on family studies, has been described as consistent with an autosomal dominant trait with reduced penetrance (Bell 1951; Hersh et al. 1953; Dutta 1965; Leung and Kao 2003). However, there have been no formal genetic association studies of fifth finger clinodactyly. Thus, the specific genes associated with the trait remain unknown. Due to its pervasiveness, mild forms of clinodactyly represent a useful target for modern gene mapping approaches, which require large samples. Furthermore, investigating the genetic basis of mild clinodactyly may offer insights on the more rare and severe form of the condition. This idea has been borne out by genetic studies of other typical-range morphological traits such as height, where discovered variants tend to be near genes implicated in rare growth disorders and syndromes (Guo et al. 2018). In the present study, we performed a genome-wide association study (GWAS) of fifth finger clinodactyly, treated as either a continuous quantitative trait or as a dichotomous trait, in three cohorts of US adults followed by meta-analysis.
2 Materials and Methods
2.1 Ethics Statement
This study was carried out in accordance with the core principles outlined in the World Medical Association Declaration of Helsinki. All participants provided written consent prior to participation in any research activities, and all study activities were approved by the University of Pittsburgh's Human Research Protection Office (protocols: STUDY19080127; STUDY19080178).
2.2 Study Samples
We included participants from three cohorts in this study. The first cohort was recruited as part of the COHRA2 study between 2011 and 2022. This was an epidemiological study designed to investigate the social, biological, and microbial determinants of oral health disparities in Appalachia (Neiswanger et al. 2015). A total of 532 unrelated participants met our inclusion/exclusion criteria: had a digital hand scan available; had genotype data; were over 18 years of age at the time of assessment; had no visible or reported hand deformities or injuries; and had no reported genetic syndromes. Because the COHRA2 study recruited mother–child pairs, only adult female participants were available. All the COHRA2 participants included in the present study were recruited from the Western Pennsylvania region. The average age of these participants was 31.1 years (range: 18.7–44.4).
The second cohort was recruited as part of the OFC2 study between 2014 and 2019. This was a study designed to investigate the genetic factors that contribute to orofacial cleft risk. A total of 99 unrelated participants (54 females; 46 males) met our inclusion/exclusion criteria: had a digital hand scan available; had genotype data; were over 18 years of age at the time of assessment; had no visible or reported hand deformities or injuries; and had no reported genetic syndromes. Thirteen of these participants had an isolated oral cleft; these individuals were not excluded because we showed previously that there is no evidence of association between clinodactyly and orofacial clefting (Lee et al. 2022). All the OFC2 participants included in the present study were recruited from the Western Pennsylvania region. The average age of these participants was 37 years (range: 19–80).
Participants from the COHRA2 and OFC2 cohorts were also included in an earlier phenotypic study looking at the effects of sex, age, and body size on fifth finger clinodactyly (Lee et al. 2022). This earlier study did not include any genetic analysis.
The third cohort included participants recruited as part of the GABC study between 2006 and 2009 (Desch et al. 2013). This study was focused on identifying genetic factors involved in blood clotting and recruited healthy individuals from the University of Michigan undergraduate and graduate population as well as their biological siblings. As part of the study's phenotypic assessment, data on a series of common morphological traits was captured, including fifth finger clinodactyly. To meet the study's listed inclusion criteria individuals had to be between 14 and 35 years of age, “generally healthy” with at least one eligible sibling, and had to have an active University of Michigan email address. The exclusion criteria listed were “acute or chronic disease requiring regular visits to a physician,” a history of a bleeding or blood-clotting disorder, and pregnancy. GABC data was obtained through dbGaP (accession: phs000304.v3.p1, Accessed on April 12, 2023). The dbGaP dataset includes 1151 individuals representing 503 sibships. Eight participants were either missing data on clinodactyly or relevant covariates, leaving 1143 eligible participants with usable phenotypic data. An additional 28 participants did not have genotype data, leaving a final dataset of 1115 (696 females; 419 males) available for genetic analysis. The average age of these participants was 21.1 years.
2.3 Phenotyping Approach
Participants in the COHRA2 and OFC2 studies had high-resolution digital hand scans collected. Measurements of fifth finger clinodactyly were then obtained from the scans. As described previously in Lee et al. (2022), “[p]articipants were asked to place their hands palm down on a large format flat-bed scanner (Epson GT-20000, Nagano, Japan). Participants were instructed to place their hands on the glass with light pressure and their fingers extended and separated in a natural, unstrained posture. The digital scans were imported into the program tpsDIG2 (http://sbmorphometrics.org/index.html) and landmarks were placed on the fifth finger at the midpoint of the metacarpophalangeal crease, the distal interphalangeal joint, and the distal margin of the fingertip. Using x, y coordinates from the collected landmarks, the angle of the distal interphalangeal joint (between the middle and distal phalanges) was calculated to capture the degree of fifth finger radial curvature.” Earlier studies have shown low measurement error (ICCs > 0.97) associated with landmark-based measurements of fingers from these same digital scans (Gremba and Weinberg 2018). Angles were obtained on both the right and left fifth fingers and averaged. If the angle was only available on one hand, the individual was dropped from the analysis. For COHRA2, 11 participants were dropped for the quantitative trait analysis due to missing data on one hand, leaving a final sample size of 521 available for this analysis. The mean clinodactyly angles were 3.43 (sd = 1.52) and 3.51 (sd = 1.78) degrees for the COHRA2 and OFC2 cohorts, respectively. Examples of individuals with low and high angles are shown in Figure 1.
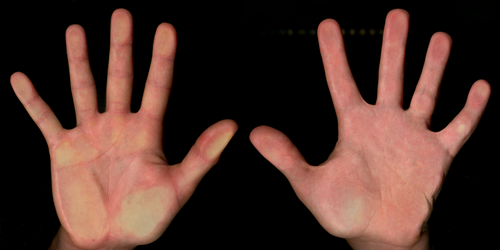
Participants in the GABC cohort provided self-reports of their fifth finger curvature status through an online survey. Participants were asked “Does the tip of your little finger (5th digit) bend toward your other fingers?” This was accompanied by a picture showing an example of the trait. Thus, clinodactyly in the GABC dataset was operationalized as a simple binary outcome: present or absent. Of the 1143 GABC participants with available phenotypes, 115 (10%) indicated that clinodactyly was present. The difference in phenotyping presented a challenge for meta-analysis across the cohorts. To solve this, we re-phenotyped our COHRA2 cohort to convert quantitative clinodactyly into a binary trait. This was accomplished by ranking the COHRA2 participants by degree of curvature and then applying a 10th percentile threshold (matching GABC) to identify the subset with clinodactyly under the new binary definition. When a COHRA2 participant's measurements were available for both hands, we used the side with the largest angle for ranking. If the measurement was only available for one hand, then this information was used for ranking. Our thresholding strategy resulted in 54 instances of fifth finger clinodactyly for the COHRA2 cohort.
Thus, for two cohorts (COHRA2 and OFC2) clinodactyly was defined as a quantitative continuous variable, and for two cohorts (GABC and COHRA2) clinodactyly was alternatively defined as a binary trait.
2.4 Genotyping
For the COHRA2 cohort, DNA samples were obtained using Oragene saliva kits (DNA Genotek Inc., Ontario, Canada). Participants were genotyped with the Illumina Infinium Multi-Ethnic Global-8 v1.0 array with genome build GRCh37/hg19, which consists of 1,742,710 genotyped single nucleotide polymorphisms (SNPs) and 5540 intensity-only probes. Genotypes were lifted over to build GRCh38 and imputed using the TOPMed Imputation Server (https://topmed.nhlbi.nih.gov/). Standard data cleaning and imputation pipelines were implemented. Imputed genotypes with a certainty above 0.9 were included for variants that had high imputation quality (R2 > 0.5) and a minor allele frequency (MAF) of at least 5%, resulting in 6,484,508 SNPs available for analysis.
For the OFC2 cohort, DNA samples were obtained using Oragene saliva kits (DNA Genotek Inc., Ontario, Canada). Participants were genotyped with the Infinium Global Diversity Array-8 v1.0 Array with genome build GRCh37/hg19, which consists of 1,894,665 genotyped SNPs and 9934 intensity only probes. Genotypes were lifted over to build GRCh38 and imputed using the TOPMed Imputation Server. Imputed genotypes with a certainty above 0.9 were included for variants that had high imputation quality (R2 > 0.5) and a MAF of at least 5%, resulting in 6,525,160 SNPs available for analysis.
For the GABC cohort, DNA samples were obtained from peripheral blood. Genotyping was performed on the Illumina Human Omni 1 M Quad v1_B with genome build GRCh36/hg18, which consists of 979,854 genotyped SNPs and 36,569 intensity-only probes. Genotypes were imputed using the 1000 Genomes cosmopolitan reference panel. Imputed genotypes with a certainty above 0.9 were included for variants that had high imputation quality (dosage R2 > 0.5), MAF of at least 5%, and lifting over to build GRCh38, resulting in 4,296,526 SNPs available for analysis.
2.5 GWAS and Meta-Analysis
We conducted GWASs for the quantitative phenotype in the COHRA2 and OFC2 cohorts. For both cohorts, PLINK (Purcell et al. 2007) was used to test for association as a linear regression model. We then conducted a second round of GWASs for the binary phenotype in the GABC and COHRA2 cohorts. For COHRA2, PLINK was used to test for association as a logistic regression model. For GABC, the GENESIS package in R (Gogarten et al. 2019) was used to perform a mixed-model GWAS to account for the relatedness among participants.
For each GWAS, covariates included age and calculated genetic ancestry PCs. We calculated ancestry PCs using KING (Manichaikul et al. 2010). For COHRA2 and OFC2, we included a total of six ancestry PCs. For GABC, we included four ancestry PCs. Due to the presence of related individuals in the GABC cohort, we calculated ancestry PCs of unrelated subjects and then projected these onto related subjects. The relevant PC plots are shown in Figure S1. We did not include sex as a covariate because prior work by our group showed no difference between males and females in the degree of fifth finger curvature (Lee et al. 2022).
We then performed two separate meta-analyses combining the results of the quantitative trait GWASs (COHRA2 and OFC2) and the binary trait GWASs (GABC and COHRA2). We conducted the meta-analyses in METAL (Willer et al. 2010) using Stouffer's p value-based meta-analysis method. There were 6,089,133 overlapping SNPs between COHRA2 and OFC2 and 3,913,557 overlapping SNPs between GABC and COHRA2.
For all analyses, we used the conventional p-value threshold of p < 5 × 10−8 to declare genome-wide statistical significance and a more liberal p-value threshold of p < 1 × 10−6 to identify potential suggestive associations.
3 Results
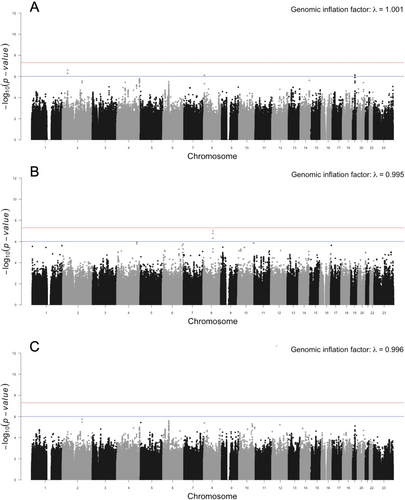
For the quantitative phenotype, we did not identify any genome-wide significant loci in either the COHRA2 or OFC2 cohorts. We did find suggestive loci on chromosomes 2, 8, and 19 in COHRA2, but these did not replicate in OFC2. The lead SNPs at these three loci are shown in Table 1. The full list of suggestive SNPs is provided in Table S1. We did not find any candidate genes at these three loci with direct relevance to clinodactyly. The quantitative trait meta-analysis did not reveal any additional loci of interest. Manhattan plots for the quantitative trait analyses are provided in Figure 2.
| Quantitative phenotype | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| COHRA2 | OFC2 | Meta-analysis | |||||||||
| Chr | SNP (BP) | N | Effect allele (freq) | beta | p | N | Effect allele (freq) | beta | p | N | p |
| 2 |
rs78276682 (41091302) |
521 | C (0.06) | 1.034 | 2 .53 × 10 −7 | 99 | C (0.07) | −0.667 | 1.73 × 10−1 | 620 | 2.90 × 10−5 |
| 8 |
rs6558883 (4437659) |
521 | T (0.26) | 0.540 | 7.86 × 10 −7 | 99 | T (0.17) | 0.076 | 8.33 × 10−1 | 620 | 3.99 × 10−6 |
| 19 |
rs78351749 (38978608) |
521 | A (0.17) | −0.616 | 7.43 × 10 −7 | 99 | A (0.26) | 0.053 | 8.58 × 10−1 | 620 | 7.98 × 10−6 |
| Binary phenotype | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| COHRA2 | GABC | Meta-analysis | |||||||||
| Chr | SNP (BP) | N | Effect allele (freq) | beta | p | N | Effect allele (freq) | beta | p | N | p |
| 9 |
rs6477432 (9972348) |
532 | A (0.12) | −0.275 | 4.11 × 10−1 | 1087 | A (0.18) | 0.984 | 7.90 × 10 −7 | 1619 | 3.51 × 10−4 |
| 9 |
rs10118945 (77039041) |
532 | C (0.08) | 1.403 | 9.58 × 10 −7 | 1115 | C (0.07) | 0.544 | 5.82 × 10−2 | 1647 | 1.40 × 10−5 |
| 16 |
rs12921393 (7147387) |
532 | G (0.31) | 0.000 | 9.98 × 10−1 | 1102 | G (0.32) | 0.804 | 4.21 × 10 −7 | 1634 | 3.24 × 10−5 |
| 20 |
rs6056500 (9260191) |
532 | C (0.05) | 0.310 | 4.95 × 10−1 | 1115 | C (0.10) | 1.277 | 4.93 × 10 −7 | 1647 | 6.01 × 10−6 |
- Note: Bold values indicate p-value less than 1 × 10–6.
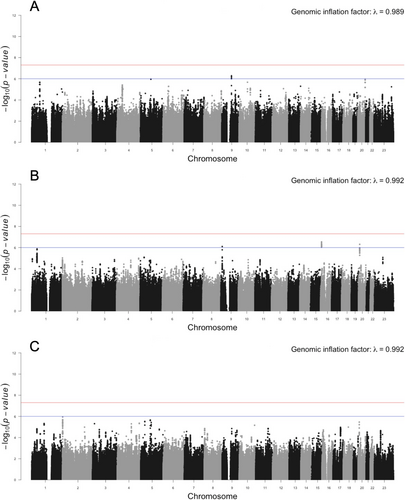
Likewise, for the binary phenotype, we did not identify any genome-wide significant loci in either the GABC or COHRA2 cohorts. For GABC, we found suggestive associations on chromosomes 9, 16, and 20. For COHRA2, there was also a suggestive hit on chromosome 9, but in a different region than the association in GABC. The lead SNPs at these three loci are shown in Table 1. We did not find any candidate genes at these four loci with direct relevance for clinodactyly. However, the candidate gene PLCB4 (OMIM: 600810) on chromosome 20 has been reported in cases of auriculocondylar syndrome (OMIM: 602483; Bukowska-Olech et al. 2021) which includes camptodactyly (OMIM: 114200) as a feature (Aponte et al. 2022). The binary trait meta-analysis did not reveal any additional loci of interest. Moreover, when we compared the COHRA2 suggestive associations between the quantitative and binary phenotypes—same cohort, different phenotype definitions—we found no consistency in the signals, suggesting that they are likely false positives. Manhattan plots for the binary trait analyses are provided in Figure 3.
For GABC, where we had sufficient sample size, we also ran an additional GWAS with the MAF threshold lowered to 1% to capture more of the genome. This analysis also failed to reveal any genome-wide significant associations (Figure S2). Those results, along with associated QQ and LocusZoom plots for the aforementioned GWASs, are provided in Figures S3–S5.
4 Discussion
Overall, our GWASs failed to detect any replicable associations for fifth finger clinodactyly. This was the case regardless of whether we defined the trait as a measured angle or a binary outcome. Instead, we observed a pattern of cohort-specific suggestive associations, more indicative of false-positive findings. These negative results should not be interpreted as definitive evidence that there is no genetic basis for fifth finger clinodactyly. While that is a possibility, our modest cohorts lack the power to detect variants with very small phenotypic effects, which are expected to drive a large proportion of heritable variation in common morphological traits (Gibson 2012).
The absence of any substantial and replicable GWAS signals does at least suggest that a major gene acting in an autosomal dominant manner is likely not involved. Even with the small sample sizes here, such an effect would likely be evident. One explanation may be that mild “typical-range” clinodactyly and severe “clinically actionable” clinodactyly represent distinct entities with distinct etiologies. Supporting the idea of a mild–severe distinction in etiology, we previously showed in 1295 adults that mild clinodactyly (measured from hand scans, as in the present study) does not exhibit the same sex bias and symmetrical presentation that is often reported in more severe forms (Lee et al. 2022). The samples used in this earlier study overlap with the ones used here.
In prior studies looking at familial inheritance patterns (Bell 1951; Hersh et al. 1953; Dutta 1965; Leung and Kao 2003), individuals were recruited from patient populations seeking treatment for clinodactyly; the majority of these cases would be on the severe side of the phenotypic spectrum. In contrast, in the present study, we assessed clinodactyly in unselected cohorts, each recruited for other primary purposes. Severe clinodactyly would be expected to be depleted in these cohorts. In fact, from our prior study of phenotypic patterns involving two of these same adult cohorts (Lee et al. 2022), we observed severe clinodactyly (defined as a deviation greater than 10%) in less than 1% of cases. Therefore, while it cannot be ruled out that more severe forms of clinodactyly will still show a Mendelian pattern (this remains to be tested), what is clear from our study is that the more common, mild form of clinodactyly looks more multigenic or potentially non-genetic in origin.
There were additional limitations beyond the small size of our cohorts. We were not able to obtain quantitative phenotypes on the GABC cohort due to the manner of assessment, preventing single quantitative trait meta-analysis across the three cohorts. Another downside to forcing traits with continuous variation into a dichotomized outcome is the resulting loss of information. Likewise, the self-reported traits in the GABC cohort are likely to have a high degree of subjectivity, especially when the trait in question is morphological and shows continuous variation. This can result in phenotype specification errors. These factors can reduce statistical power in GWAS. However, this limitation can be overcome to some degree with very large sample sizes, as shown in studies using self-reported traits in large Biobanks (Eriksson et al. 2010). Repeating these analyses with larger sample sizes may yield improved results, although we are currently unaware of other cohorts with genome-wide markers and fifth finger clinodactyly information available. It is feasible to overcome this limitation because clinodactyly is an externally visible trait that can be collected and measured on a large scale from images of the hand, whether obtained through digital flatbed scanning (such as in the current study) or simple photography.
Author Contributions
Myoung Keun Lee: data curation, formal analysis, visualization, writing – original draft preparation. Noah Herrick: investigation, writing – review and editing. Mary L. Marazita: funding acquisition, resources, writing – review and editing. John R. Shaffer: project administration, writing – review and editing. Seth M. Weinberg: conceptualization, funding acquisition, project administration, resources, writing – original draft preparation.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
Phenotype data for COHRA2 and OFC2 participants is available through the NIDCR's FaceBase controlled-access data repository at accession numbers FB00001366 and FB00001368. Genotype data for these datasets are available through dbGaP at accession numbers phs001591.v2.p1 and phs002815.v2.p1. Genotype and phenotype data for GABC participants are available through dbGaP at accession number phs000304.v3.p1.




