Atypical features and de novo heterozygous mutations in two siblings with Cockayne syndrome
Abstract
Background
Cockayne syndrome (CS) is a rare autosomal recessive disorder which displays multiorgan dysfunction, especially within the nervous system including psychomotor retardation, cerebral atrophy, microcephaly, cognitive dysfunction, mental retardation, and seizures. Many genetic variations reported were related to this syndrome, but splicing mutations with cardiac anomalies have not been found in previous studies.
Methods
Herein, we described a pair of brothers and sisters who present essential manifestations of CS including premature feature, developmental delay, growth failure, microcephaly, and characteristic facial features, such as sunken eyes and a beaked nose. Interestingly, the brother also presented with atypical features which included cardiac anomalies such as left atrioventricular enlargement and cardiac dysfunction such as dilated cardiomyopathy. In addition, whole exome sequencing and RNA sequencing were employed to analyze their genetic landscape.
Results
WES analysis showed that these two cases carried double unreported heterozygous spliced mutations in the excision repair cross-complementing group 8 (ERCC8, also known as CSA, NM_000082) gene, which were c.78-2 (IVS1) A>T and c.1042-1 (IVS10) G>A, respectively. Moreover, transcript sequencing analysis validated these mutation sites. In this study, Gene Ontology enrichment and KEGG pathway analyses from RNA sequencing demonstrated similarities but some differences when compared with previous studies.
Conclusion
For patients with Cockayne syndrome, cardiac changes need to be monitored carefully, especially for cases with splicing mutations of the ERCC8 gene.
1 INTRODUCTION
Cockayne syndrome (CS; MIM # 133540, 216400) was first identified by Cockayne EA in 1932 (Cockayne, 1936), and was initially a series of clinical cases with different mutation sites (Bertola et al., 2006; Sanchez-Roman et al., 2018; Taghdiri et al., 2017). The characteristic features of this disease include progressive neurodegeneration, growth restriction, hearing loss, skin photosensitivity, premature aging, and eye anomalies (Karikkineth, Scheibye-Knudsen, Fivenson, Croteau, & Bohr, 2017). This disease resulted from a gene mutation in either of the ERCC8 (CSA) or ERCC6 (CSB) genes-encoding proteins involved in transcription-coupled repair (TC-NER), such as DNA repair, or transcription and chromatin remodeling (Fousteri, Vermeulen, van Zeeland, & Mullenders, 2006). Although the severity of CSA (MIM # 216400) and CSB (MIM # 133540) patients is different, their phenotypes mostly overlap. The ERCC8 gene encodes a Walker domain repeat protein, which interacts with CS type B (CSB) protein and with p44 protein, a subunit of the RNA polymerase II transcription factor IIH (Fischer et al., 2011; Fousteri et al., 2006). The cells from CS patients are abnormally sensitive to ultraviolet radiation and are defective in the repair of transcriptionally active genes in vitro and in vivo. Several transcript variants encoding different isoforms have been found for this gene. In this study we revealed two de novo spliced mutation sites that contribute to the pathogenesis of CS group A (MIM # 216400). Further transcript sequencing between case 2 and the parent may give further insights into the nature of this syndrome. These results will help to explain the molecular pathogenesis of this disease and provide new clues whereby future targeted treatment may be developed.
2 MATERIALS AND METHODS
2.1 Ethical compliance
Our study was approved by the Research Ethics Committee of Children's Hospital of Soochow University. All research procedures were conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from the family involved.
2.2 Gene sequencing analysis
Peripheral blood from the patients and their parents were subjected to whole exonic mutation analysis and these procedures were the same as in our previous report (Kong et al., 2019). According to genotype–phenotype analysis, we ultimately obtained putative mutations. Sanger sequencing was performed to verify the splicing mutations found by whole exome sequencing. For chr5: 60224788-60224788c.78-2 (IVS1) A>T, we amplified a 436 bp fragment with an annealing temperature set at 55 ºC. The primer sequences were as follows: F: 5'-AGCAGGTTTTACTGCATTTCAC-3' and R: 5'-TGTCCATTAGATGTCCTCAGCTAC-3'. Whereas for chr5:60183348-60183348c.1042-1 (IVS10) G>A, we amplified a fragment of 1040 bp in size at an annealing temperature of 60°C using primers F: 5'-GGCCTATTTTAGTAGACGCTCA-3' and R: 5'-GTTTACAATAATCAACAGAAATGCCT-3'. Copy number variant (CNV) analysis was used and sequenced according to Illumina's NovaSeq6000 platform standardized process. Raw data were derived from the Illumina official basecall analysis software BclToFastq. Next, copy CNV of sequences of 100 kb and above were detected, filtered, and annotated. Lastly causative variants were found by CNV hazard rating. RAN sequencing was performed following the manufacturer's protocol and the data were analyzed according to previous reports (Kim, Langmead, & Salzberg, 2015). Bioinformatic analysis methodology employed in this study is described in the Methods part of the supplemental data section; the data of proband (WQL) were normalized to that of healthy parents.
3 RESULTS
3.1 Clinical report
3.1.1 Case 1 (proband, SQL)
A 5-year-old male subject was admitted to hospital with tachypnea which had lasted for 2 weeks and a cough for 2 days. Previous history showed ventricular enlargement at 1 year of age and reduction in cerebral myelination and cerebellar atrophy at 2 years, based on head nuclear magnetic resonance imaging. At 8 months preterm, a fetal heart monitor indicated that the baby had suffered from anoxia for some time, but the symptoms disappeared after oxygen therapy. Eventually the baby was born by cesarean section for post-term pregnancy. The main features presented were initially developmental delays which became progressive and obvious after the age of 1 year. On admission to hospital at 8 years and 3 months of age, his weight was 10 kg (< −3 SD), height was 95 cm (< −3 SD), and head circumference 46 cm (< −3 SD). Upon physical examination, the following were observed: skin pigmentation, subcutaneous fat loss, microcephaly, enophthalmia, bird-shaped nose, tachypnea, enamel hypoplasia, hip joint contracture, hip abduction restriction, and undescended testis, which were verified on both sides by ultrasound tests (Figure 1a). Lung computed tomography showed that inflammation occurred on both lungs, and the left main bronchus was compressed, and the heart was enlarged (Figure 1b). Cardiac examination revealed muffled heart sounds and further cardiac color ultrasound indicated left atrioventricular enlargement and left ventricular contractile function was induced while blood pressure was within the normal range (Figure 1c).
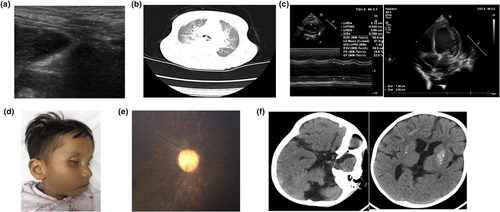
3.1.2 Case 2 (proband's sister, WQL)
The case was admitted to hospital after vomiting and with fever for 1 day and with seizures for half a day at the age of 2 years and 7 months. Previous history showed she presented with intrauterine growth retardation and cesarean delivery was implemented at 34 + 1 weeks of pregnancy. She was then sent to hospital for further care as her birth weight was too low (1,800 g). She experienced mental, physical, and language developmental delay. The primary features of this patient were developmental delay such as enamel hypoplasia, mental retardation, and a thin body. On admission, her body weight was 7,500 g (< −3 SD), height 76 cm (< −3 SD), and head circumference 43 cm (< −3 SD). Physical examination findings resembled those of her brother except for tachypnea, joint contracture, enamel hypoplasia, and normal genitals (Figure 1d). Further fundus examination showed abnormal retinal pigment and fine retinal vessels indicating retinal atrophy (Figure 1e). Only persistent left superior vena cava was found and no deterioration in cardiac function was seen with ultrasonography. The cardiac picture of their mother was normal. Calcification can be seen in the both globus pallidus and the subcortical white matter using cerebral computed tomography (CT) scan; sulci widening, dilated ventricles, and shrunken cerebellar hemisphere were also seen (Figure 1f).
3.2 Molecular analysis
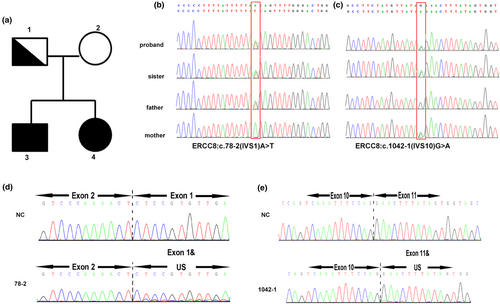
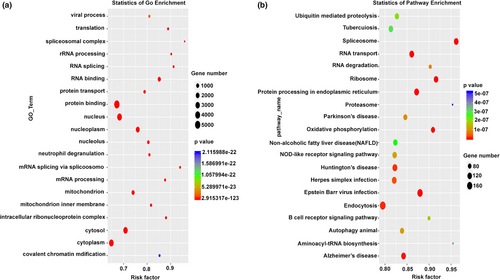
To further confirm the diagnosis and understand the genetic background, whole exome sequencing (WES) and CNV-seq were performed. WES data analysis indicated that the two cases presented were double heterozygous in the ERCC8 gene, which represents c.78-2 (IVS1) A>T and c.1042-1 (IVS10) G>A, respectively. Both of these are novel splice sites, and the father was found to be the carrier of c.78-2(IVS1) A>T and the mother the carrier of c.1042-1 (IVS10) G>A. CNV-seq was also carried out to identify the likely causative gene, but no pathogenic CNV was identified in either pair of siblings. To further validate the spliced mutation site, RNA sequencing was employed to analyzed WQL and the patient (Figure 2). Further Gene Ontology (GO) enrichment indicated a biological process including regulation of transcription, DNA-templated transcription, and DNA-templated transport. The oxidation-reduction process, protein ubiquitination, cellular response to DNA damage stimulus, and mitochondrial inner membrane were highlighted. Many cellular components such as membrane, nucleus, cytoplasm, and mitochondria relating genes were also found. Protein binding–related genes were the most common (Figure 3a). In order to determine the major biochemical and signal transduction pathways, a pathway enrichment analysis was performed by KEGG enrichment. A total of 20 enriched pathways were measured in this analysis, many common pathways relevant to this disorder were revealed in our study (ubiquitin-mediated proteolysis, spliceosome, RNA transport, RNA degradation, Ribosomal, protein processing within the endoplasmic reticulum, oxidative phosphorylation, endocytosis, and aminoacyl-tRNA biosynthesis, Figure 3b). The specific upregulated and downregulated genes seen between the case and her parents are presented in the Table S1.
4 DISCUSSION
The prevalence of CS is 2.5 per million live births (Kubota et al., 2015). Despite the low morbidity of this disease, it has poor prognosis with no drug treatments available or cure. Recently, symptomatic treatment and avoidance of sunlight have been the only way to increase life span. Diverse origins in the pathogenesis of CS can cause a range in phenotype severity, depending on the mutation pattern and other genetic and nongenetic factors resulting in multiple organ degeneration and death. In this study, the siblings presented very similar clinical manifestation of CS type I. They have classical characteristics including nervous system degeneration such as calcification of the globus pallidus and subcortical white matter, sulci widening, dilated ventricles, cerebellar atrophy, microcephaly, as well as cognitive decline, developmental delay, enamel hypoplasia, premature facial features, retinal atrophy, joint contracture, skin pigmentation, and progressive loss of body fat. The symptoms are progressive and typically become apparent after 1 year of age. The presentation of our patients is consistent with most reported cases of the disease (Karikkineth et al., 2017). However, the sister showed intrauterine growth retardation which is mostly displayed in type II CS involving the ERCC6 gene mutation, indicating that clinical manifestations can overlap in the different subtypes. The proband (brother) manifested left atrioventricular enlargement and a decrease in left ventricular ejection fraction which to our knowledge has never been reported in this disease previously. This manifestation may be another feature of CS. The symptom relating to cardiovascular structural dysfunction has rarely been seen, except for two studies (Ovaert, Cano, & Chabrol, 2007; Pasquier et al., 2006). The mechanism by which this phenomenon occurs is elusive. One hypothesis is that abnormal RNA synthesis might affect the aortic wall.
Whole exome, CNV, and transcript sequencing were performed to understand the pathogenesis and genomic background of CS. Using these techniques, we found two de novo splice mutations in the ERCC8 gene leading to deficient RNA transcription, or other mechanisms responsible for the many clinical observations. As an important member of the NER repair pathway, ERCC8 is necessary for transcription-coupled nucleotide excision repair recognition. However, in CS patients, many of the clinical observations are different from other NER defective disorders, suggesting that other factors should be considered also to account for this variation. Furthermore, GO enrichment of transcript data not only revealed genes involved in biological processes and cellular components which are mostly linked to transcription repair systems but also showed gene anomalies within the mitochondria and mitochondrial inner membrane. Mitochondrial abnormalities have been found in the cells of CS patients previously (Cleaver et al., 2014). Mitochondrial changes consisting of increased mitochondrial membrane potential, superoxide production, and increased oxygen consumption rates were observed in a CSA deficiency model in vivo and in vitro, caused by stalled ribosomal DNA transcription and subsequent PARP1 activation (Scheibye-Knudsen et al., 2016).
Therefore, CSA nuclear mutations cause various biological manifestations, in particular, cardiac changes, especially in our case. We found marked changes in protein binding–related genes which may be an alternative explanation for the clinical manifestations seen. In order to find the most salient biochemical and signal transduction pathways, we used pathway enrichment analyses and found ubiquitin-mediated proteolysis and transcription-couple nucleotide excision repair pathways, which have also been seen by others (Karikkineth et al., 2017; Sommers, Suhasini, & Brosh, 2015). We also found many novel pathways which require further experimentation for their elucidation.
ACKNOWLEDGMENTS
This work was supported by fund from Suzhou Science and Technology Development Project (SYS 201757), the Natural Science Foundation of Jiangsu Higher Education Institutions of China (18KJB320022), and National Natural Science Foundation of China (project code 81571551).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




