Daurisoline suppresses esophageal squamous cell carcinoma growth in vitro and in vivo by targeting MEK1/2 kinase
Donghao Wang, Weizhe Zhang, and Xiaofan Zhang contributed equally to this article.
Abstract
Esophageal squamous cell carcinoma (ESCC) accounts for 90% of esophageal cancers and has a high mortality rate worldwide. The 5-year survival rate of ESCC patients in developing countries is <20%. Hence, there is an urgent need for developing new and effective treatments that are based on newly-discovered emerging molecules and pathways to prevent ESCC occurrence and recurrence. We investigated the effects of Daurisoline, a bis-benzylisoquinoline alkaloid extracted from the rhizome of menisperum dauricum, on ESCC cell proliferation and elucidated the molecular mechanisms underlying its functions. To explore the effects of Daurisoline on ESCC growth in vitro and in vivo, cell proliferation assays and anchorage-independent growth assays were performed and a patient-derived xenograft (PDX) model was established. Subsequently, phosphoproteomics, molecular docking analysis, pull down assays, mutation experiments and in vitro kinase assay were performed to explore the mechanism of Daurisoline's function on ESCC. Daurisoline inhibited ESCC proliferation in vitro and reduced ESCC PDX exnograft growth in vivo by reducing ERK1/2 phosphorylation. Furthermore, it directly bound to MEK1 (at Asn78 and Lys97) and MEK2 (at Asp194 and Asp212) kinases to inactivate the ERK1/2 signaling pathway. Our results suggest that Daurisoline is a dual inhibitor of MEK1 and MEK2 and suppresses ESCC growth both in vitro and in vivo by inactivating the ERK1/2 signaling pathway. This is first report on the use of MEK inhibitor for ESCC and highlights its potential applications for ESCC treatment and prevention.
Abbreviations
-
- EAC
-
- esophageal adenocarcinoma
-
- EC
-
- esophageal cancer
-
- ESCC
-
- esophageal squamous cell carcinoma
-
- GSEA
-
- gene set enrichment analysis
-
- KEGG
-
- Kyoto Encyclopedia of genes and genomes
-
- LC-MS/MS
-
- liquid chromatography tandem mass spectrometry
-
- PDX
-
- patient-derived xenograft
-
- SCID
-
- severe combined immunodeficiency
-
- SHEE
-
- Shantou human embryonic esophageal
-
- TCGA
-
- The Cancer Genome Atlas
1 INTRODUCTION
Esophageal cancer (EC) ranks seventh and sixth in incidence and mortality worldwidely among all cancers, respectively.1 Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma are two main pathological classifications of EC.2 A total of 90% of EC cases worldwide are reported as ESCC.3 Clinical treatments of ESCC include mainly surgery, chemotherapy and radiotherapy, however, the 5-year survival rate of ESCC patients is poor (<20%).4-6 Therefore, it is of great clinical significance to improve treatments with effective therapeutic outcomes that prevent the occurrence and recurrence of ESCC.
The mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signal pathway plays an essential role in the proliferation, differentiation, and metastasis of cancer cells.7, 8 This indicates that MEK1/2 kinases play the pivotal roles in malignant transformation and maybe rational therapeutic targets in cancers. Most MEK1/2 inhibitors have been developed and applied in a clinical setting.9, 10 For example, Selumetinib is used to treat melanoma, and thyroid cancer as well as plexus neurofibroma.11, 12 Nevertheless, MEK inhibitors currently have limited clinical use for ESCC treatment.
An effective strategy of discovering new antitumor drugs is to screen natural compounds. Daurisoline, bis-benzylisoquinoline alkaloid, is a traditional chinese medicine and extracted from the rhizome of menisperum dauricum.13 Accumulating studies demonstrate that Daurisoline has effective and pharmacological effects on a serious of diseases, such as platelet aggregationreper, focal ischemia and thrombotin A2 inhibition.14-18 Daurisoline is reported to inhibit the cell proliferation of lung cancer and hepatocellular carcinoma.19, 20 This indicates Dasurioline has the potential to become a promising therapeutic compound for cancers.
Here, we attempted to investigate the effects of Daurisoline on ESCC cell proliferation in vitro and patient-derived xenograft (PDX) growth in vivo, particularly to elucidate the molecular mechanisms underlying functions of Daurisoline by phosphoproteomics, computer docking models, pull down assays and in vitro kinase assays.
2 MATERIALS AND METHODS
2.1 Compound and antibodies
Daurisoline (CAS:70553-76-3) was purchased from Shanghai Liding Biotechnology Co., Ltd., (Shanghai, China; purity ≥98%, Cat No. DR10996). Antibodies to detect MEK1/2 (#12671S), ERK1/2 (#9125S), p-ERK1/2 T202/Y204 (#94370S), MEK1(#2352S), MEK2 (#9125S) and p-MEK1/2 (#2338S) were purchased from CST, Ki67 (ab16667), p62 (sc-28359) and LC3-α/β (sc-398822) antibodies were obtained from Abcam, β-tubulin antibody(ET1602-4) from Hua An Biotechnology Co., Ltd., and glyceraldehyde 3-phosphate dehydrogenase antibody (60004-1-Ig) from ProteinTech.
2.2 Cell culture
Shantou human embryonic esophageal (SHEE) cell lines was a gift from prof. Enmin Li of Shantou University; ESCC cell lines (KYSE150 and KYSE450) were purchased from the Cell Bank of the Chinese Academy of Sciences. The cells were cytogenetically verified by STR-Promega and authenticated. Cells were cultured in RPMI-1640 medium (Biological Industries), 10% fetal bovine serum and incubated in a 1:1 mixture of 1% antibiotics. The culture condition for all cells was 37°C with 5% CO2.
2.3 Cytotoxicity assay
A total of 8000 cells/well of KYSE150, KYSE450, and SHEE cells were seeded in 96-well plates and cultured for 12–16 h. Then the culture medium was replaced with growth medium containing different doses of Daurisoline (0, 1, 2.5, 5, 10, 20, and 40 μM). All cells were fixed in 4% paraformaldehyde after culturing for 0, 24, or 48 h. The cells were stained with DAPI and counted by In Cell 6000 Analyzer. The curves of cell numbers were plotted with various concentrations of Daurisoline at different times. Each point represents the mean ± standard deviation (SD) of three biological replicates.
2.4 Cell proliferation assays
KYSE150 (1000 cells/well) and KYSE450 (1000 cells/well) cells were seeded in 96-well plates and cultured for 12–16 h. Then the culture medium was replaced with growth medium containing different doses of Daurisoline (0, 2.5, 5, 10, and 20 μM). All cells were fixed in 4% paraformaldehyde after culturing for 0, 24, 48, 72, or 96 h. The cells were stained with DAPI and counted by In Cell 6000 Analyzer. The curves of cell numbers were plotted with various concentrations of Daurisoline at different times. Each point represents the mean ± SD of three biological replicates.
2.5 Anchorage-independent growth assays
The 8000 cells of KYSE150 and KYSE450 were seeded in the medium supplemented with 0.3% agar and different concentrations of Daurisoline (0, 2.5, 5, 10, and 20 μM) in 6-well plates. A base layer contained 0.6% agar with Daurisoline. The cells were cultured at 37°C and 5% CO2 for 10 days, and the number of colonies was counted by IN Cell Analyzer 6000.
2.6 Anchorage-dependent growth assays
KYSE150 and KYSE450 cells (300 cells/well) were treated with 20 μM Daurisoline in six-well plate. After culturing for 2 weeks, the cell colonies were dyed by crystal violet, and were photographed and counted.
2.7 LC-MS/MS analysis
After Daurisoline treatment for 24 h, KYSE150 cells were in lysis buffer (8 M urea, 1% Protease Inhibitor Cocktail) and sonicated by a high intensity ultrasonic processor (Scientz) (Note: For PTM experiments, inhibitors were also adCBd to the lysis buffer, e.g., 3 μM TSA and 50 mM NAM for acetylation.). The remaining CBbris was removed by centrifugation at 12,000g at 4°C for 10 min. Finally, the supernatant was collected and concentrated with BCA kit.
The tryptic peptiCBs were dissolved in 0.1% formic acid (solvent A) (Full lane elution), directly loaCBd onto a home-maCB reversed-phase analytical column (15 cm length, 75 μm i.D.). The gradient was increase from 6% to 23% solvent B (0. 1% formic acid in 98% acetonitrile) over 26 min, 23%–35% in 8 min and climbing to 80% in 3 min then holding at 80% for the last 3 min, all at a constant flow rate of 400 nl/min on an EASY-nLC 1000 UPLC system.
The MS/MS results were processed by Maxquant search engine. A total of 20 ppm was set in first search and 5 ppm was in main for the mass tolerance for precursor ions, and the mass tolerance for fragment ions was set as 0. 02 Da. False Discovery Rate (FDR) was adjusted to <1% and minimum score for modified peptiCBs was set >40. To test the enrichment of the differentially modified protein, encyclopedia of Kyoto Encyclopedia of Genes and Genomes (KEGG) database were used to iCBntify enriched pathways by a two-tailed Fisher's test. A p < 0.05 was significant. Specific experimental methods were conducted according to our previous research.21, 22
2.8 Western blotting
Proteins were obtained from KYSE150 and KYSE450 cell lysates (Solarbio). The concentration of cellular protein was measured with the BCA protein assay kit (Beyotime). The protein samples were electrophoresed by 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to a polyvinylidene fluoride (PVDF) membrane (BIO-RAD). The membranes were blocked with 5% skimmed milk at room temperature (RT) for 2 h, then the primary antibodies were incubated overnight at 4°C. At the next day, these membranes were incubated with horseradish peroxidase (HRP)-immunoglobulin G antibody at RT for 2 h, and were detected by eBlot Touch Imager (Shanghai Yibuote Optoelectronic Technology Co., Ltd.).
2.9 Spearman's correlation analysis
The expression level of the MEK1/2 gene in EC was analyzed using of the UALCAN databases (http://ualcan.path.uab.edu/analysis.html). The map of two-gene correlation and multigene correlation were caculated by ggstatsplot (R package) (https://github.com/Indrajeet Patil/ggstatsplot/), and pheatmap (R package) (https://cran.r-project.org/web/packages/pheatmap/), respectively. A p < 0.05 denoted statistical significance.
2.10 Molecular docking model between Daurisoline and MEK1/2
First, we downloaded the 2D structure of Daurisoline (PubChem CID: 51106) from PubChem (https://pubchem.ncbi.nlm.nih.gov). Then, the structural models of MEK1 (PDB:3EQH) and MEK2 (PDB:1S9I) were downloaded from the Protein Data Bank (PDB) database (https://www.rcsb.org/). All the small molecule ligands and water molecules were deleted, then the hydrogen atoms were added. The docking models between Daurisoline and MEK1/2 were completed using AutoDock 4.2.6 software (http://autodock.scripps.edu/) under default parameters. Finally, the models were visualized by PyMOL (The PyMOL Molecular Graphics System, Version 2.3.4).
2.11 Pull down assays
The following experimental procedures were performed according to the instructions provided by GE Healthcare Biosciences. First, Sepharose 4B dry powder (0.3 g) was activated with 1 mM HCl. The activated Sepharose 4B beads and the compound Daurisoline (3 mg) were added to the 40% dimethyl sulfoxide (DMSO)/H2O (vol/vol) coupling solution and rotated overnight at 4°C. The mixture was then transferred to 0.1 M Tris-HCl buffer (pH 8.0) and rotated again overnight at 4°C. The beads were washed 3 times with 0.1 M acetic acid buffer containing 0.5 M NaCl (pH 4.0). The beads were then washed three times with 0.1 M Tris-HCl (pH 8.0) containing 0.5 M NaCl. Daurisoline-conjugated Sepharose 4B Beads or Sepharose 4B beads were obtained. Finally, active MEK1/2 proteins (250 ng) or KYSE150/450 cell lysates (500 μg) were added to the reactor and rotated overnight at 4°C. The beads were washed 3 times with washing solution, and the proteins bound to the beads were boiled and analyzed by Western blotting.23
2.12 MEK1/2 mutation and overexpression in 293F cells
The sequences of wild-type (WT) MEK1 and MEK2 were amplified from KYSE450 complementary DNA (cDNA) by PCR and inserted to pLVX-IRES-3xflag vector (Cat No. VT8006; YouBia Biotechnology Company) by XhoI/NotI sites. The mutational sites of MEK1 and MEK2 protein were showed as follow: N78A (Asn78 to Ala) and K97A (Lys97 to Ala) for MEK1 protein; D194A (Asp194 to Ala) and D212A (Asp212 to Ala) for MEK2 protein. The mutated PCR productions were obtained by nested PCR and were cloned to XhoI/NotI sites of pLVX-IRES-3xflag vector. The primer sequences of MEK1 and MEK2 WT and mutation protein were showed in Table 1. These wild-type and mutant plasmids were transfered into 293F cells. After incubating for 72 h, the MEK1/2-overexpressed cells were harvested and lysated by sonication for the pull down assays.
| Gene name | Type | Primer sequence (5′–3′) |
|---|---|---|
| MEK1 | WT Forward: | TATTCTCGAGATGCCCAAGAAGAAGCCGACG |
| WT Reverse: | TATGCGGCCGCgGACGCCAGCAGCATGGGTTG | |
| N78A Forward: | TCAGTGAGCTGGGGGCTGGCGCCGGCGGTGTGGTGTTCAAGGT | |
| N78A Reverse: | ACCTTGAACACCACACCGCCGGCGCCAGCCCCCAGCTCACTGA | |
| K97A Forward: | CTGGCCTGGTCATGGCCAGAGCCCTAATTCATCTGGAGATCAA | |
| K97A Reverse: | TTGATCTCCAGATGAATTAGGGCTCTGGCCATGACCAGGCCAG | |
| MEK2 | WT Forward: | TATTCTCGAGATGCTGGCCCGGAGGAAGCCG |
| WT Reverse: | TATTCTCGAGATGCTGGCCCGGAGGAAGCCG | |
| D194A Forward: | AGCACCAGATCATGCACCGAGCCGTGAAGCCCTCCAACATCCT | |
| D194A Reverse: | AGGATGTTGGAGGGCTTCACGGCTCGGTGCATGATCTGGTGCT | |
| D212A Forward: | GAGGGGAGATCAAGCTGTGTGCCTTCGGGGTGAGCGGCCAGCT | |
| D212A Reverse: | AGCTGGCCGCTCACCCCGAAGGCACACAGCTTGATCTCCCCTC |
2.13 Cellular thermal shift assay
A total of 20 μM Daurisoline was added and treated KYSE450 cells for 4 h. The cultured cells were harvested and resuspended with phosphate buffered saline (PBS). The cell lysates were divided into 100 μl aliquot part and treated at different temperatures for 3 min, respectively. Cell lysates were frozen twice with liquid nitrogen. The lysate was separated from the cell debris by centrifugation at 12,000g for 20 min at 4°C. The supernatants were transferred to the new microtubes and analyzed by Western blotting.
2.14 In vitro kinase assay
The active MEK1 (100 ng) or MEK2 (50 ng) kinase was mixed with different concentrations of Daurisoline or vehicle (DMSO) at RT for 15 min. Then, the inactive ERK2 protein (250 ng) and ATP (100 μM) were added and incubated at 30°C for 30 min. Finally, the phospholated proteins were analyzed by Western blotting.
2.15 Gene set enrichment analysis (GSEA)
To observe the expression difference between the Daurisoline treated and control groups, the phosphoproteome datasets were analyzed by the GSEA V4.0.3 (http://www.broadinstitute.org/gsea/). Further, details were listed in the references.22
2.16 Knockdown of ERK2 by short hairpin RNA (shRNA)
We inserted shRNA for human ERK2 (Forward primer: 5′-CCGGAGCCAGGATACAGATCTTAACTCGAGTTAAGATCTGTATCCTGGCTTTTTTG-3′, Reverse primer: 5′-AATTCAAAAAAGCCAGGATACAGATCTTAACTCGAGTTAAGATCTGTATCCTGGCT-3′) into the pLKO.1 vector. We established the shERK2 stable ESCC celll lines (KYSE150 and KYSE450) by transfecting with short hairpin RNA and verified the transfection efficiency by Western blotting.
2.17 CRISPR/Cas9 knockout cell lines
To knockout the ERK2 gene in KYSE150 cells by using the CRISPR/Cas9 system, the lentiCRISPRv2-sgERK2 plasmids were contructed. Following the manufacturer's suggested protocols, sgERK2 plasmids were transfected into HEK293T cells using Jet Primer (Thermo Fisher Scientific). The KYSE150 cells were infected with 8 μg/ml polybrene and viral particles and selected with 2 μg/ml puromycin for 72 h. Knockout efficiency was verified by Western blotting. The oligonucleotide sequences of ERK2 single guide (sg) RNA were designed in an online CRISPR tool (https://chopchop.cbu.uib.no) and were listed as Table 2.
| Gene name | Type | Primer sequence (5′–3′) |
|---|---|---|
| sgERK2 1 | Forward: | CACCGCAACCTCTCGTACATCGGCG |
| Reverse: | AAACCGCCGATGTACGAGAGGTTGC | |
| sgERK2 2 | Forward: | CACCGCTACTGCCAGAGAACCCTGA |
| Reverse: | AAACTCAGGGTTCTCTGGCAGTAGC |
2.18 Establishment of the ESCC PDX models
Female severe combined immune deficency (SCID) mice (Vital River Labs, Beijing, China) aged 6–8 weeks were used as PDX models. The sample was EG20 (ESCC, male, T2N0M0, moderately differentiated, obtained from Linzhou Cancer Hospital, Henan Province, China). The patient was fully informed of the study and provided consent. The tumors were divided into chunks of about 0.1 g and planted subcutaneously into the backs of mice. Mice were randomly divided into the three groups: control group, 20 and 40 mg/kg Daurisoline group. Daurisoline or DMSO was administered daily by intraperitoneal injection. Tumor volumes and weights were measured every 3 days. A month later, the mice were euthanized and the tumors were extracted. The tumor volume (mm3) was calculated according to the following formula (length × width × width/2). This study was approved by the Ethics Committee of Zhengzhou University (ZZUHCI-2019012). More experimental details were reported in our previous methods.24
2.19 Immunohistochemical staining
The molecular changes in PDX model were detected by immunochemical staining. Tumors were embedded in paraffin and slides were dewaxed and hydrated with xylene and gradient alcohols. The tissues were treated with epitologic repair and soaked in 0.3% hydrogen peroxide for 5 min. Subsequently, the slides were incubated with antibodies (Ki67, p-MEK1/2 S221, and p-ERK1/2 T185/Y187) at 4°C overnight. After incubation with HRP secondary antibody at 37°C for 15 min, the slides were detected by 2, 4-diaminobenzidine substrate (DAB) and hematoxylin. TissueFAXS was used to scan all the slides and analyze them by HistoQuest 4. 0 software (TissueGnostics GmbH). The complete experiment was found in our previous instructions.24
2.20 Statistical analyses
For data statistical analysis, SPSS 24.0 software (IBM) was applied in this experiment, and the results were expressed as the mean ± SD. The difference between groups was calculated by student t-test, one-way analysis of varianc or Kruskal–Wallis H-test. The experiments were repeated three times, and p < 0.05 indicated statistically significant difference.
3 RESULTS
3.1 Daurisoline inhibited ESCC cell proliferation in vitro
Daurisoline (50 μM) was cytotoxic to KYSE450 cells through screening the compounds from the nature compound bank of Shanghai Sunny Biotech Co., Ltd by cytotoxicity assays (Figure 1A and Table 3). Daurisoline is a bis-benzylisoquinoline alkaloid (Figure 1B). After Daurisoline treatment for 24 and 48 h, the half-maximal inhibitory concentrations (IC50) of SHEE, KYSE150, and KYSE450 cells were 76.26, 20.73, and 13.56 μM at 24 h, and 33.50, 11.02, and 9.94 μM at 48 h, respectively (Figure 1C). After Daurisoline treatment (0, 2.5, 5, 10, 20 μM), the cell numbers were 10952, 10498, 9219, 8350, 6610, and 10940, 9524, 8033, 4881, 3351 in KYSE150 and KYSE450 at 72 h, respectively. And the cell numbers were increased to 21495, 20111, 19635, 16491, 11928 for KYSE150 and 19812, 19051, 17660, 11355, 6176 for KYSE450 at 96 h, p < 0.05 (Figure 1D). These results indicated that Daurisoline inhibited the proliferation of KYSE150 and KYSE450 cells in a dose-dependent manner. Similarly, the clone numbers of KYSE150 and KYSE450 cells were 2033, 1632, 1452, 921, 615 and 2278, 1774, 1541, 1175, 965 after Daurisoline treatment in anchorage-independent growth assay, respectively (Figure 1E). These results indicated that Daurisoline effectively inhibited ESCC cell proliferation in vitro.
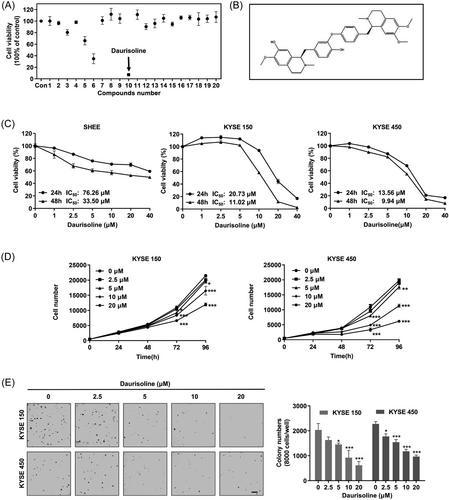
| Drug serial number | Drugs name |
|---|---|
| 1 | Kumatakenin |
| 2 | Loganetin |
| 3 | 3′-hydroxy Puerarin |
| 4 | 23-Hydroxybetulinic acid |
| 5 | β-ecdysone |
| 6 | Cycloastragenol |
| 7 | Emodin-1-O-glucoside |
| 8 | Desmethoxyyangonin |
| 9 | Daphnoretin |
| 10 | Daurisoline |
| 11 | EsculentosideA |
| 12 | Fraxin |
| 13 | Geniposidic acid |
| 14 | Alantolactone |
| 15 | Homoplantaginin |
| 16 | 1-(3,4-Dimethoxycinnamoyl)piperidine |
| 17 | Isovanillic acid |
| 18 | l-Cysteine hydrochloride anhydrous |
| 19 | PiperlotineC |
| 20 | Chrysosplenetin |
3.2 Phosphorylation of ERK1/2 was downregulated after Daurisoline treatment in ESCC
To investigate the molecular mechanism underlying the inhibition of Daurisoline, the quantitative phosphoproteomics was applied. We treated KYSE150 cells with 20 μM Daurisoline for 24 h (Figure 2A). The first-order mass error of most spectrograms was within 10 ppm, which conformed to the high precision characteristic of orbital well mass spectra (Figure S1A). Most peptides were 7–20 amino acids in length. The distribution of the peptide lengths identified conformed to the quality control requirements by mass spectra (Figure S1B). The numbers of secondary spectra obtained were 383,273. After screening, the numbers of available effective spectrograms were 94,520. The 12,965 phosphorylation sites were identified on 3130 proteins, of which 7415 sites on 2500 proteins contained quantitative data. To ensure a high credibility of the results, localization probability > 0.75 was used to filter the identification data. Consequently, we identified 8549 sites on 2861 proteins, of which 6601 sites on 2408 proteins included quantitative data (Figure S1C). The protein quantification group was normalized to remove the influence of protein expression on the modified signal. The screening criteria for different sites were as follows: 1.5 times the change threshold and two-sample two-tailed t-test (p < 0.05). Based on these data and standards, we found that the phosphorylation of 176 sites was enhanced and that of 340 sites was reduced in the Daurisoline treated cells (Figure S1D and Table S1). The raw mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD036419.
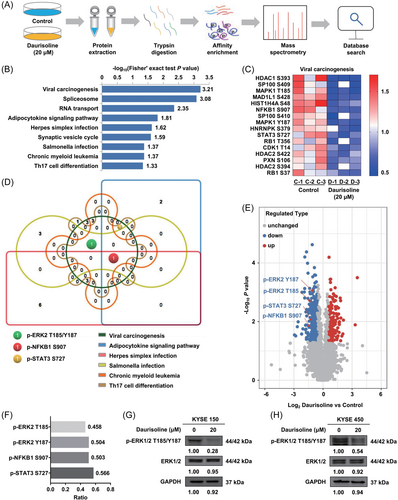
A total of 340 downregulated phosphorylation sites were enriched in KEGG pathway. The KEGG pathway was selected according to Fisher's exact test (p < 0.05). Ultimately, nine KEGG pathways were screened: viral carcinogenesis, spliceosome, RNA transport, adipocytokine signaling pathway, herpes simplex virus infection, synaptic vesicle cycle, salmonella infection, chronic myeloid leukemia, and Th17 cell differentiation (Figure 2B). Most of the phosphorylation sites were downregulated from the heat map results which showed the levels of the phosphorylation sites in the viral carcinogenesis pathway (Figure 2C). In six KEGG pathways, multiple proteins were repeated in the different pathways. Key proteins in these pathways were found by the Wayne diagram basing on the six KEGG pathways (Figure 2D). The NFκB1 protein was all enriched in six pathways, the ERK2 protein in four pathways, and the STAT3 protein in three pathways. The original mass spectra of p-ERK2 T185/Y187 were shown in the Figure S1E. In the volcanic map, according to the −log10 (p-value), the four phosphorylation sites were ranked from top to bottom as p-ERK2 Y187 (3.018), p-ERK2 T185 (2.498), p-STAT3 S727 (1.743), and p-NFκB1 S907 (1.569) (Figure 2E). In addition, the ratios of the above four phosphorylation sites were 0.504, 0.458, 0.566, and 0.503, respectively (Figure 2F). We speculated that p-ERK2 T185/Y187 were important phosphorylation sites in the above nine KEGG pathways. In addition, although ERK1 was not enriched in the KEGG pathway, the phosphorylation of ERK1 was also downregulated in the phosphoproteomics data. The level of p-ERK2 T185/Y187 was also significantly decreased after Daurisoline (20 μM) treatment for 24 h in KYSE150 (Figure 2G) and KYSE450 cells (Figure 2H). These results indicated that Daurisoline inhibited the phosphorylation of ERK1/2 in ESCC.
3.3 Daurisoline could directly bind with MEK1 and MEK2 kinase
To evaluate the target of Daurisoline, we predicted upstream kinases for p-ERK2 T185 and p-ERK2 Y187 in the above nine KEGG pathways by using IGPS1.0 software. MEK1-7 (MAP2K1-7), and ALK and LTK were found as the upstream kinase proteins of p-ERK2 T185 and p-ERK2 Y187 (Table S2), respectively. To verify these, we performed the pull down assays (Figure S2). The protein binding with Daurisoline was analyzed by MS. MEK1, MEK2, MEK3, and MEK7 were considered as the binding protein according to MS results (Table S3). ERK1/2 was the sole substrate of MEK1/2 kinase, p38 MAPK and JNKs were the substrates of MEK3 and MEK7, respectively.25 Therefore, combining above phosphoproteome data, we deduced that Daurisoline downregulated ERK2 phosphorylation by targeting MEK1/MEK2 kinase.
MEK1 and MEK2 poteins were highly expressed in ESCC.26 Additionally, the data from the Cancer Genome Atlas (TCGA) database also revealed that MEK1 (Figure S3A) and MEK2 (Figure S3B) were both significantly overexpressed in a variety of cancers, including ESCA (Figure S3C,D). Then, we conducted a multi-gene correlation by analysising TCGA ESCC data. We observed that there was a significant correlation between MEK1 and ERK2 in ESCC (Figure S3E,F). These data indicated that MEK1/2 kinase maybe a potential target for ESCC.
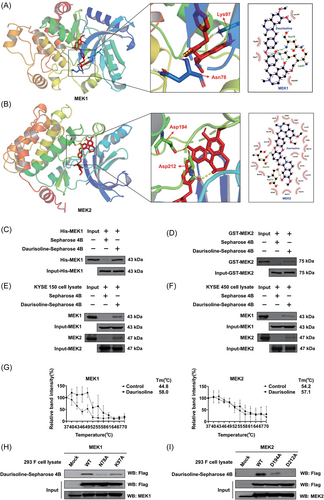
AutoDock models indicated that Daurisoline could bind with MEK1 kinase (binding energy: −11.06 kcal/mol) and MEK2 kinase (binding energy: −8.50 kcal/mol). Asn78 and Lys97 of MEK1, and Asp194 and Asp212 of MEK2 were hydrogen bond with Daurisoline (Figure 3A,B). Pull down assays displayed that Daurisoline could not only directly bind to recombinant MEK1 and MEK2 protein (Figure 3C,D), but also bind to MEK1 and MEK2 protein kinase in KYSE150 and KYSE450 cell lysate (Figure 3E,F). What's more, to demonstrate Daurisoline directly binds to MEK1 and MEK2 kinase in intact cells, we applied the cell thermal shift assay. The Tm for MEK1 was increased from 44.8°C to 58°C in Daurisoline-treated cell, and MEK2 was increased from 54.2°C to 57.1°C (Figure 3G and Figure S3G). These results indicated that Daurisoline bound to the MEK1 and MEK2 protein directly in the intact cells. Furthermore, to verify the accuracy of the binding sites of MEK1/2 kinase, we mutated the afore mentioned amino acids of MEK1 (Asn78 and Lys97) and MEK2 (Asp194 and Asp212) with alanine (Ala, A). We expressed the mutational proteins in 293F cells, then applied the pull down assays to certify the binding between Daurisoline and mutational MEK1 protein. The results showed that the binding ability of Daurisoline to the MEK1 N87A and MEK1 K97A protein was significantly reduced, compared with MEK1 WT protein (Figure 3H). Interestingly, after Asp194 and Asp212 mutation, the binding efficiency between Daurisoline and mutational MEK2 protein was also significantly reduced (Figure 3I). This data indicated that Daurisoline is a novel dual MEK1 and MEK2 inihibitor.
3.4 Daurisoline inhibited the MEK1/2-ERK1/2 signaling pathway in ESCC
We found the phosphorylation level of ERK2 was in turn reduced at 2.5, 5, 10, and 20 μM Daurisoline by in vitro kinase assay (Figure 4A,B). These results suggested Daurisoline could cause a decrease in MEK1/2 kinase activity and directly leaded to suppress ERK2 activation. ERK1/2 protein was enriched and significantly downregulated in the ERBB signaling pathway (Figure 4C) and prostate cancer pathway (Figure 4D) by GSEA analysis. In addition, MEK2 (MAP2K2) was also enriched in the afore mentioned two pathways. We proved that Daurisoline inhibited ERK1/2 phosphorylation in KYSE150 and KYSE450 cells and had no significant effect on MEK1/2 phosphorylation (Figure 4E,F). We also checked the protein levels of p62 and LC3-α/β which are the autophagy markers and found that Daurisoline did not change the levels of p62 or LC3-α/β after Daurisoline treatment in KYSE150 (Figure S4A) and KYSE450 (Figure S4B) cells.

Furthermore, to certify whether Daurisoline-induced anticancer effects can be bothered after silencing ERK2, we constructed the ERK2 knockdown KYSE150 and KYSE450 cell lines by transfecting with short hairpin RNA (Figure 5A) and found that knocking down ERK2 could inhibit the cell proliferation (Figure 5B) and clony formation (Figure 5C) in KYSE150 and KYSE450. After Daurisoline treatment, the proliferation of KYSE150 shERK2 and KYSE450 shERK2 cells was not inhibited (Figure 5D) and the clone formation ability (Figure 5E and Figure S5A) was not decreased. Followly, we knocked out ERK2 gene in KYSE150 cells by CRISPR/CAS9 system (Figure 5F). Our results demonstrated that knocking out ERK2 could inhibit the cell proliferation (Figure 5G) and clony formation (Figure 5H) of KYSE150. As excepted, the proliferation (Figure 5I) and clony formation (Figure 5J and Figure S5B) of sgERK2 KYSE150 was not inhibited after Daurisoline treatment. This data indicated that Daurisoline-induced anticancer effects could be bothered after silencing ERK2. Therefore, these above results indicated that Daurisoline downregulated the MEK1/2-ERK1/2 signaling pathway in ESCC cells.
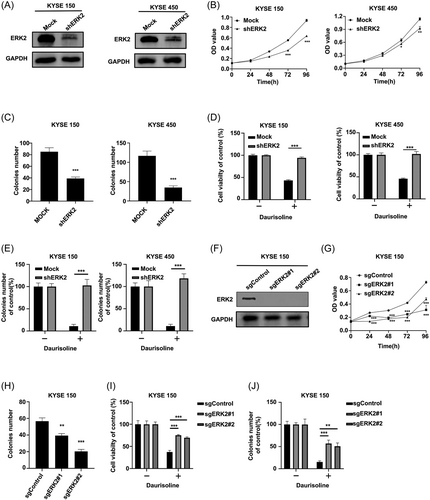
3.5 Daurisoline inhibited ESCC PDX tumor growth
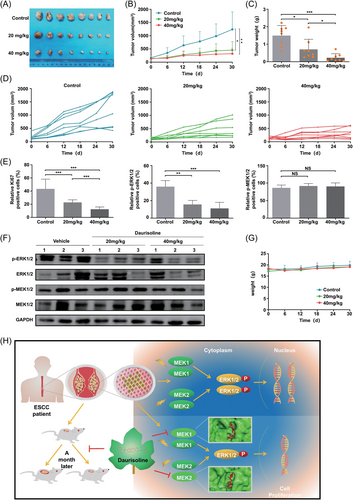
To investigate the anticancer effect of Daurisoline in vivo, we established ESCC PDX models of ESCC (Figure 6A). After 30 days, the final tumor volumes were 1238.4 ± 660.20, 454.07 ± 322.20, 320.486 ± 201.60 mm3 in control, 20 and 40 mg/kg Daurisoline-treated group, respectively (Figure 6B). We found that the tumor weights also decreased with the increasing dosages of Daurisoline (Figure 6C) and the growth inhibition rates were 52.77% and 84.74% in the 20 and 40 mg/kg Daurisoline-treated groups compared with control group. The tumor growth curves of each mouse in Daurisoline-treated group and control group were showed to verify (Figure 6D). The results of HIC showed that the postive rates of the Ki67 and phospho-ERK1/2 were decreased and the positive rate of p-MEK1/2 did not change significantly after Daurisoline treatment (Figure 6E and Figure S6). The results of Western blotting displayed that p-ERK1/2 was decreased in Daurisoline-treated group (Figure 6F), which were consistent with the HIC results. Interetingly, there were no significant differences in the body weights between Daurisoline-treated group and the control group (Figure 6G), indicating that Daurisoline had less toxic or side effects. In conclusion, Daurisoline suppressed ESCC tumor proliferation by MEK1/2-ERK1/2 signal pathway in vivo.
4 DISCUSSION
It is of great clinical significance to screen drugs that prevent the development and recurrence of ESCC.6 Traditional chinese medicine has been used in clinical settings and compounds extracted from traditional chinese medicine represent a promising resource for anticancer drug development.27 By screening natural compounds, we identified that Daurisoline exhibited anticancer effects on the proliferation of ESCC cells. Additionally, the PDX models retain the histological and genetic characteristics of the original tumors; and are more convincing than the cell experiments.28, 29 Daurisoline displayed a significant antitumor effect on ESCC PDX growth in vivo (Figure 6H). Thus, our study demonstrates that Daurisoline could be an attractive therapeutic agent for ESCC treatment.
MAPK cascades are key signaling pathways that contribute to cancer cell proliferation, survival and metastasis.30 Dysfunction in the MEK-ERK pathway is a major trigger for the development of most cancer types.8, 31 Mutations in RAS or RAF gene typically promote malignancies by over-activating MEK and ERK downstream.12 MEK1 and MEK2 proteins are the kinases, and activation of the MEK/ERK cascade has been reported in ESCC. Inhibition of MEK1/2 specifically shuts off ERK1/2 signaling without directly affecting other signaling pathways.26 Thus, we checked the activating of ERK1/2 signaling by protein phosphorylation in ESCC. Our results identified that Daurisoline bound directly with MEK1 and MEK2 proteins (Figure 3) and further inhibited the kinase activity of MEK1 and MEK2 by in vitro kinase assay (Figure 4A,B). Followly, the protein and protein phosphorylation level of ERK1/2 were both examined (Figure 4E,F). The results demostrated that only protein phosphorylation level of ERK2 were decreased, the total protein level of ERK2 was not changed after Daurisoline treatment. It is also proved that the inhibitary function of Daurisoline was aborted after downregulating ERK2 protein (Figure 5). Therefore, Daurisoline suppresses the cell proliferation of ESCC by MEK-ERK signal pathway.
Development of drugs targeting the inhibition of MEK is an attractive strategy for the treatment of cancers. Several MEK inhibitors have demonstrated anticancer activity in patients with NSCLC.32 In our study, phosphoproteome data confirmed that Daurisoline inhibited ERK1/2 phosphorylation in ESCC cells (Figure 2), molecular docking and pull down data (Figures 3 and 4) showed that Daurisoline directly bound to both MEK1 and MEK2 protein kinases. MEK1/2, as the targets of tumor therapy, are highly selective and unique.32, 33 MEK1 and MEK2 share 80% similarity in the amino acid sequence of the kinase domain.34 Most MEK1/2 inhibitors are ATP noncompetitive inhibitors such as allosteric inhibitors. The deep cleft located between the small and large lobes is the catalytic site of MEK1/2 kinase.35 The lysines (Lys97/101 of MEK1/2) in the β3 strand couple α- and β-phosphates of ATP, which facilitates the formation of a salt-bridge between the lysine and a conserved glutamate (Glu114/118 in MEK1/2) near the center of αC-helix in the small lobe.9 In our study, Daurisoline binds with Lys97 of MEK1 (Figure 3A), this indicated that Daurisoline competes with ATP to bind with MEK1 and is a type I kinase inhibitor, constituting the majority of ATP-competitive inhibitors for MEK1. Nevertherless, Daurisoline interacts with the Asp194 and Asp212 sites of MEK2 kinase (Figure 3B), indicating it is an ATP-noncompetitive allosteric inhibitor for MEK2. Of note, daurisoline suppresses lung cancer tumorigenesis by targeting HSP90 directly.19 In our study, daurisoline directly binds to MEK1 and MEK2 in KYSE150 and KYSE450 cells by CETSA assay and inhibits the ERK2 phosphorylation in vitro and in vivo. Therefore, Daurisoline is a dual MEK1 and MEK2 inhibitor to shut off ERK signaling pathway in ESCC.
Daurisoline has been reported to inhibit cancer growth by interrupting autophagy.20, 36 To identify whether Daurisoline inhibited the autophagy in ESCC, we checked the protein level of autophagy markers (p62 and LC3-α/β) in ESCC cells after Daurisoline treatment and found Daurisoline could not inhibited the autophagy in ESCC. In ESCC, most studies have described that activation of autophagy promotes ESCC carcinogenesis by the AMPK/ULK1 or mTORC1/ULK1 or RPTOR/ULK signaling pathways.37, 38 There is also no report about the relationship between MEK/ERK signal pathway and autophagy in ESCC. Consequently, we consider that Daurisoline didn't inhibit the autophagy by MEK/ERK signal pathway in ESCC.
In summary, the present study evaluates Daurisoline as a dual MEK1 and MEK2 inhibitor inactivates aberrant ERK signaling pathway in ESCC. Moreover, Daurisoline exhibits a high ESCC cell-killing activity by simultaneously inhibiting ERK phosphorylation. This is the first report of an MEK inhibitor that has an effective impact on ESCC, as currently, no MEK inhibitors have been applied in the clinical treatment of ESCC. Therefore, Daurisoline has potential to confer significant therapeutic benefits for ESCC patients in the future.
AUTHOR CONTRIBUTIONS
Donghao Wang and Yanan Jiang: wrote the original manuscript. Weizhe Zhang, Xiaofan Zhang, and Qiong Wu: conducted cell experiments. Mingzhu Li and Weizhe Zhang: performed site-directed mutation experiments on amino acids. Lili Zhao: carried out the immunohistochemical staining experiment. Qiang Yuan and Yin Yu: extracted the purified protein. Jing Lu and Jimin Zhao: analyzed the data. Yanan Jiang and Xin Li: revised the paper. Zigang Dong, Kangdong Liu, and Yanan Jiang: directed the design of the project. All authors have read and approved the final manuscript.
ACKNOWLEDGMENTS
Thanks to Bo Li, Xining Liu, Yubing Zhou, Zhuo Bao, Yaxing Wei, Xiangyu Wu, and Ang Li from the China US (Henan) Hormel Cancer Institute for their help. This research was supported by the National Natural Science Foundations of China (No. 81872335; No. 32000650), National Natural Science Youth Foundation (No. 81902486), National Science & Technology Major Project Key New Drug Creation and Manufacturing Program, China (No. 2018ZX09711002), The Central Plains Science and Technology Innovation Leading Talents (NO. 224200510015), Science and Technology Project of Henan Province (No. 212102310187), Zhengzhou University of Scientific Research Fund (32213272).
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The datasets during and/or analysed during the current study available from the corresponding author on reasonable request.