Donor polymorphisms of toll-like receptor 4 associated with graft failure in liver transplant recipients†
This work was supported by the National Institutes of Health Genomics of Transplantation program through an American Recovery and Reinvestment Act supplement (5U19-AI070119).
Abstract
There have been many reports showing significant associations between recipient genetic variants and allograft outcomes, including acute rejection and graft failure, but less is known about the contribution of the donor genotype. We analyzed 37 single-nucleotide polymorphisms (SNPs) within the toll-like receptor 4 (TLR4) gene from deceased donor liver allografts transplanted into 738 recipients to determine their effects on liver graft failure (LGF). Two SNPs exhibited a significant association with LGF after adjustments for donor race and recipient race and corrections for multiple test comparisons: rs11536865 [hazard ratio (HR) = 2.5, P = 0.0003] and rs5030717 (HR = 1.67, P = 0.0008). An additional SNP, rs913930, exhibited a significant association in Caucasian donors (HR = 1.62, P = 0.0006), and 2 SNPs exhibited a suggestive association in African American donors: rs11536865 (HR = 2.45, P = 0.002) and rs5030717 (HR = 2.32, P = 0.002). Additionally, the liver donor risk index (HR = 2.56, 95% confidence interval = 1.54-4.26, P = 0.0003) and the recipient hepatitis C virus (HCV) status (HR = 1.53, 95% confidence interval = 1.04-2.24, P = 0.032) increased the risk of all-cause LGF in a Cox proportional hazards model adjusted for recipient race. Donor polymorphisms in TLR4 could be important factors in modulating TLR4 activity and, therefore, affect the risk of graft loss. Additionally, there is a suggestion of an interaction between polymorphisms within TLR4 and the HCV status. Liver Transpl, 2012. © 2012 AASLD.
There have been many attempts to associate solid organ allograft outcomes with specific genetic variants, usually in the form of single-nucleotide polymorphisms (SNPs). Most studies have focused on recipient genotypes, but there is evidence that donor-associated genetic variants also play a role in affecting allograft outcomes. For example, donor variation in the nuclear pregnane X receptor (nuclear receptor 1I2) has been associated with delayed graft function after renal transplantation,1 and donor variants of mannose binding lectin and donor heme oxygenase 1 have been associated with liver allograft outcomes, including graft survival.2, 3
Several recipient SNPs within the toll-like receptor 4 (TLR4) gene have been reported to be associated with kidney allograft dysfunction.4, 5 Additionally, variants in TLR4 have been associated with ischemia and reperfusion injury in liver allografts, and TLR4 signaling has been associated with acute rejection in the liver.6, 7 The TLR4 gene product plays a critical role in activating immune responses to bacterial infections. TLR4 is responsible for recognizing lipopolysaccharide from gram-negative bacteria and inducing an inflammatory response resulting in the production of several proinflammatory, antiviral, and antibacterial cytokines.8, 9 In a healthy liver, the expression level of TLRs is low, but in the presence of lipopolysaccharide or a liver injury (eg, alcoholic liver disease or cirrhosis), the expression of TLR4 increases.9-12 It has been shown that increased TLR4 expression in the liver is associated with hepatic inflammation and liver injury.13, 14 Increased TLR4 expression has also been hypothesized to be associated with acute rejection in liver allografts.7, 15 Increased TLR4 expression in kidney allografts has also been associated with acute rejection and chronic allograft nephropathy.16 The presence of the CC genotype of TLR4 promoter SNP rs10759932 in either the recipient or the donor has been associated with rejection-free survival in kidney transplantation.5 Two common coding polymorphisms within the extracellular domain, rs4986790 (p.Asp299Gly) and rs4986791 (p.Thr399Ile), that have been shown to attenuate tumor necrosis factor α secretion have been associated with reduced renal allograft rejection.17-19 The incidence of acute rejection in kidney allograft recipients was significantly reduced when the donor genotypes were heterozygous for either variant.20
We have previously reported that the TLR4 variant p.Asp299Gly loss-of-function mutation (rs4986790) in the donor liver attenuates the rate of postischemic acute liver injury, but the relevance of donor-associated TLR4 activation to long-term liver function is unclear.4 We hypothesized that variants altering TLR4 function affect transplant outcomes and result in graft damage and eventual graft loss. We report several donor TLR4 polymorphisms that are associated with an increase in liver graft failure (LGF) in liver allograft recipients.
PATIENTS AND METHODS
Abbreviations:
BMI, body mass index; HBV, hepatitis B virus; HCV, hepatitis C virus; HR, hazard ratio; LD, linkage disequilibrium; LGF, liver graft failure; MAF, minor allele frequency; MELD, Model for End-Stage Liver Disease; OPTN, Organ Procurement and Transplantation Network; PELD, Pediatric End-Stage Liver Disease; SNP, single-nucleotide polymorphism; SRTR, Scientific Registry of Transplant Recipients; TLR, toll-like receptor; UTR, untranslated region.
Research Subjects
Seven hundred thirty-eight liver donors were identified for this analysis. Lymph nodes or blood specimens were obtained by organ procurement organizations in the United States from deceased donors whose family or next of kin gave generalized consent for research before organ and tissue donation. These organ procurement organizations included LifeSource (St. Paul, MN), LifeQuest (Gainesville, FL), New Jersey Organ & Tissue Sharing Network (New Providence, NJ), Organ Donor Center of Hawaii (Honolulu, HI), Southwest Transplant Alliance (Dallas, TX), One Legacy (Los Angeles, CA), New England Organ Bank (Waltham, MA), Lifebanc (Cleveland, OH), and Louisiana Organ Procurement Agency (Metairie, LA). This study was approved by the institutional review board of the Hennepin County Medical Center (University of Minnesota). DNA was extracted from donor tissues with general laboratory methods.
Clinical data (Table 1) were obtained from the United Network for Organ Sharing via the Scientific Registry of Transplant Recipients (SRTR). The SRTR data system includes data on all donors, wait-listed candidates, and transplant recipients in the United States [submitted by the members of the Organ Procurement and Transplantation Network (OPTN)] and has been described elsewhere.21 The Health Resources and Services Administration (US Department of Health and Human Services) provides oversight for the activities of the OPTN and SRTR contractors.
| Variable | All (n = 738) | No LGF (n = 599) | LGF (n = 139) | P Value |
|---|---|---|---|---|
| Recipient factors | ||||
| Age at transplantation (years) | 51.1 ± 15.0 | 50.6 ± 15.4 | 53.1 ± 13.4 | 0.098 |
| Male sex [n (%)] | 499 (67.6) | 401 (66.9) | 98 (70.5) | 0.5 |
| Race [n (%)] | 0.22 | |||
| African American | 72 (9.8) | 57 (9.5) | 15 (10.8) | |
| Caucasian | 551 (74.7) | 452 (75.5) | 99 (71.2) | |
| Other | 115 (15.6) | 90 (15.0) | 25 (18.0) | |
| HCV-positive [n (%)] | 269 (39.0)* | 210 (37.2)* | 59 (47.2)* | 0.038 |
| HBV core antibody–positive [n (%)] | 141 (20.5)* | 113 (20.2)* | 28 (21.7)* | 0.24 |
| Total cold ischemia time (hours) | 6.5 ± 3.3 | 6.4 ± 3.3 | 6.8 ± 3.3 | 0.54 |
| Primary cause of disease [n (%)] | 0.076 | |||
| Acute hepatic necrosis | 35 (4.7) | 29 (4.8) | 6 (4.3) | |
| HCV | 151 (20.5)* | 113 (18.9) | 38 (27.3) | |
| Alcoholic liver disease | 122 (16.5) | 106 (17.7) | 16 (11.5) | |
| Cholestatic disease | 69 (9.4)* | 61 (10.2) | 8 (5.8) | |
| Metabolic liver disease | 19 (2.6) | 17 (2.8) | 2 (1.4) | |
| Malignancy | 163 (22.1) | 134 (22.4) | 29 (20.9) | |
| Other | 179 (24.2)* | 139 (23.2) | 40 (28.8) | |
| MELD (n = 680) | 6221.6 ± 9.5 | 6221.4 ± 9.3 | 6222.2 ± 10.1 | 0.37 |
| PELD (n = 23) | 6212.4 ± 11.7 | 6211.9 ± 11.8 | 6217.5 ± 13.4 | 0.0085 |
| Donor factors | ||||
| Age at organ recovery (years) | 40.0 ± 16.8 | 39.4 ± 16.7 | 42.6 ± 17.4 | 0.05 |
| Male sex [n (%)] | 460 (62.3) | 372 (62.1) | 88 (63.3) | 0.96 |
| Race [n (%)] | 0.21 | |||
| African American | 128 (17.3) | 101 (16.9) | 27 (19.4) | |
| Caucasian | 610 (82.7) | 498 (83.1) | 112 (80.6) | |
| Height (cm) | 171.6 ± 16.4 | 171.5 ± 16.8 | 171.9 ± 14.4 | 0.96 |
| Cause of death [n (%)] | 0.15 | |||
| Anoxia | 149 (20.2) | 120 (20.0) | 29 (20.9) | |
| Cerebrovascular/stroke | 283 (38.4)* | 220 (36.7) | 63 (45.3) | |
| Head trauma | 280 (37.9) | 237 (39.6) | 43 (30.9) | |
| Central nervous system tumor | 3 (0.4) | 2 (0.3) | 1 (0.7) | |
| Other | 23 (3.1) | 20 (3.3) | 3 (2.2) | |
| Donation after cardiac death [n (%)] | 43 (5.8) | 29 (4.8) | 14 (10.1) | 0.082 |
| Liver donor risk index | 1.4 ± 0.3 | 1.3 ± 0.3 | 1.4 ± 0.3 | 0.0028 |
| Partial/split transplantation [n (%)] | 22 (3.0) | 19 (3.2) | 3 (2.2) | 0.73 |
| HCV-positive [n (%)] | 22 (3.0) | 20 (3.3) | 2 (1.4) | 0.4 |
- * The values were calculated after the exclusion of samples with missing information. MELD, Model for End-Stage Liver Disease; PELD - Pediatric End-Stage Liver Disease.
Genotyping
Forty SNPs covering the entire TLR4 region were genotyped in DNA from liver donors. Genotyped SNPs were selected by SeattleSNPs. The selection was made to ensure that all haplotype blocks within the TLR4 gene were represented in both Caucasian and African American samples. The selected SNPs were submitted to Illumina (San Diego, CA) for processing with the Illumina Assay Design Tool to create a panel of SNPs for genotyping. A total of 250 ng of genomic DNA per donor was used for Illumina SNP genotyping with the Illumina BeadArray platform according to the manufacturer's protocol. Raw hybridization intensity data processing, clustering, and genotype calling were performed with the genotyping module in the BeadStudio package (Illumina). Before genotype calling, the trimmed mean intensities were calculated from the normalized intensity values obtained for each bead type on the array via the rejection of outliers to ensure the high quality of the genotype data. Genotype calls were generated with the GenCall software incorporated into the BeadStudio package.
Statistical Analysis
To examine the relationships between recipient and donor variables and all-cause LGF, univariate Cox proportional hazards models were created. To identify genetic variants associated with LGF, we performed a Cox regression for the time to LGF for each genotyped SNP with adjustments for donor race and recipient race. A subgroup analysis was also performed with Caucasian and African American donor samples separately. SNPs were coded for the additive genetic model. A robust sandwich estimator was used to account for the correlation between transplant recipients with the same donor. The Bonferroni method was used to adjust the α level for multiple comparisons in the overall analysis and separately for each subgroup analysis. The familywise type I error rate for a 2-sided significance test was taken to be 0.05.
A multivariate model was created via backwards selection with a retention P value of 0.10. The potential covariates included the following: recipient race, hepatitis C virus (HCV) status, sex, age (linear and quadratic), and body mass index (BMI; linear and quadratic); Model for End-Stage Liver Disease (MELD)/Pediatric End-Stage Liver Disease (PELD; linear and quadratic); cold ischemia time; first transplant; primary cause of liver disease; previous abdominal surgery; portal vein thrombosis; partial/split transplantation; donor race, sex, age (linear and quadratic), and height (linear and quadratic); donation after cardiac death status; cause of donor death; and liver donor risk index (linear and quadratic). Recipient race and donor race were forced into the model at all stages in the selection procedure. The covariates selected by backwards selection were included in the Cox regression model, and the association between SNPs and the time to LGF was reexamined. All statistical analyses were conducted with SAS 9.2 (SAS Institute, Cary, NC).
RESULTS
Clinical factors associated with LGF were determined for 738 recipients who underwent transplantation between 2006 and 2010, with 139 all-cause LGFs (19%) during a median follow-up period of 13.0 months (interquartile range = 9.4-25.0 months) after transplantation. An analysis based on the specific causes of LGF could not be performed because the SRTR database does not collect detailed data on causes of graft loss and all the outcome data for this analysis came from SRTR. When the donors were stratified by race, 610 recipients were found to have received livers from Caucasian donors, and 112 experienced graft failure (18%); 128 recipients received livers from African American donors, and 27 experienced graft failure (21%). According to a univariate Cox regression, the clinical factors for recipients that were associated with LGF included an HCV-positive status (P = 0.038) and a higher PELD score (P = 0.0085), although the number of pediatric recipients was low (n = 23; Table 1). Although both the cold ischemia time and the warm ischemia time have been shown to be important variables with respect to LGF, the SRTR database does not reliably collect information on the warm ischemia time, so it was not listed separately in the Cox model. The clinical factors for donors that were significantly associated with LGF included the liver donor risk index (P = 0.0028) and the age at organ recovery (P = 0.05).
Forty donor SNPs within the TLR4 gene were analyzed in all 738 liver allograft recipients for their association with LGF. Cox proportional hazards models were used with adjustments for recipient race and donor race. Three SNPs—rs1927912, rs2737196, and rs1927910—were monoallelic in our population and were excluded from further analysis. Two SNPs exhibited a significant association with LGF after we took into account multiple comparisons (α = 0.05/37 SNPs = 0.00135) and corrected for recipient race (Table 2). First, an SNP in the 5′ promoter region of TLR4, rs11536865 (c.−728G>C), showed a significant association with LGF [hazard ratio (HR) = 2.5, P = 0.0003], although the minor allele frequency (MAF) was low (MAF = 0.02); second, an SNP in intron 2, rs5030717 (c.261-833A>G), showed a significant association with LGF (HR = 1.67, P = 0.0008). Two additional SNPs in the TLR4 gene also exhibited a modest association: rs913930 (HR = 1.48, P = 0.003), which is located at 3′ of the TLR4 coding region, and rs2770146 (HR = 1.45, P = 0.004), which is located in intron 2 (c.261-1329T>C).
| SNP | Location* | All Donor Genotypes | Caucasian Donor Genotypes | African American Donor Genotypes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P Value | Allele | MAF | HR | P Value | Allele | MAF | HR | P Value | Allele | MAF | HR | ||
| rs10759930 | 5′ (−5130) | 0.06 | T | 0.36 | 0.78 | 0.06 | T | 0.38 | 0.77 | 0.77 | T | 0.13 | 0.88 |
| rs16906053 | 5′ (−4569) | 0.27 | C | 0.03 | 0.63 | Monoallelic | T | 0 | 0.24 | C | 0.18 | 0.6 | |
| rs2737191 | 5′ (−4036) | 0.0095 | G | 0.26 | 1.41 | 0.02 | G | 0.29 | 1.41 | 0.25 | G | 0.17 | 1.51 |
| rs2770150 | 5′ (−3612) | 0.0095 | G | 0.26 | 1.41 | 0.02 | G | 0.29 | 1.41 | 0.25 | G | 0.17 | 1.51 |
| rs2737190 | 5′ (−2570) | 0.56 | G | 0.38 | 0.93 | 0.74 | G | 0.32 | 0.96 | 0.38 | A | 0.3 | 0.76 |
| rs10116253 | 5′ (−2431) | 0.79 | C | 0.27 | 1.03 | 0.77 | C | 0.26 | 0.96 | 0.29 | C | 0.31 | 1.31 |
| rs1927914 | 5′ (−2026) | 0.56 | G | 0.38 | 0.93 | 0.74 | G | 0.32 | 0.96 | 0.38 | A | 0.3 | 0.76 |
| rs10759932 | 5′ (−1607) | 0.1 | C | 0.15 | 1.28 | 0.33 | C | 0.14 | 1.2 | 0.15 | C | 0.25 | 1.51 |
| rs11536865 | 5′ (−728) | 0.0003 | C | 0.02 | 2.5 | Monoallelic | G | 0 | 0.002 | C | 0.12 | 2.45 | |
| rs7864330 | Intron 1 | 0.58 | G | 0.05 | 0.85 | 0.39 | G | 0.05 | 0.73 | 0.86 | G | 0.09 | 1.08 |
| rs1927911 | Intron 1 | 0.6 | A | 0.31 | 0.94 | 0.74 | A | 0.26 | 0.95 | 0.47 | G | 0.42 | 0.81 |
| rs1927907 | Intron 2 | 0.24 | A | 0.15 | 1.21 | 0.39 | A | 0.14 | 1.17 | 0.51 | A | 0.25 | 1.26 |
| rs10983756 | Intron 2 | 0.95 | T | 0.05 | 0.98 | 0.42 | T | 0.05 | 0.74 | 0.21 | T | 0.06 | 1.79 |
| rs2770146 | Intron 2 | 0.004 | C | 0.28 | 1.45 | 0.007 | C | 0.31 | 1.46 | 0.25 | C | 0.17 | 1.51 |
| rs5030717 | Intron 2 | 0.0008 | G | 0.11 | 1.67 | 0.07 | G | 0.1 | 1.44 | 0.002 | G | 0.2 | 2.32 |
| rs5030710 | p.105Ser/Ser | 0.49 | C | 0.02 | 0.14 | Monoallelic | T | 0 | 0.05 | C | 0.11 | 0.14 | |
| rs4986790 | p.Asp299Gly | 0.43 | G | 0.05 | 0.78 | 0.58 | G | 0.05 | 0.82 | 0.68 | G | 0.08 | 0.76 |
| rs4986791 | p.Thr399Ile | 0.8 | T | 0.04 | 0.92 | 0.74 | T | 0.05 | 0.9 | 0.71 | T | 0.01 | 1.55 |
| rs5030718 | p.Glu474Lys | 0.77 | A | 0.01 | 1.24 | Monoallelic | G | 0 | 0.81 | A | 0.03 | 1.2 | |
| rs7869402 | 3′ UTR (1106) | 0.45 | T | 0.06 | 0.81 | 0.19 | T | 0.04 | 0.56 | 0.99 | T | 0.22 | 1 |
| rs11536889 | 3′ UTR (1205) | 0.05 | C | 0.14 | 0.67 | 0.06 | C | 0.16 | 0.67 | 0.6 | C | 0.04 | 0.7 |
| rs7873784 | 3′ UTR (2010) | 0.25 | C | 0.15 | 0.82 | 0.49 | C | 0.15 | 0.89 | 0.32 | C | 0.14 | 0.55 |
| rs11536897 | −3084 | 0.44 | A | 0.05 | 0.79 | 0.2 | A | 0.05 | 0.67 | 0.59 | A | 0.03 | 1.43 |
| rs1927906 | −3189 | 0.92 | C | 0.13 | 1.02 | 0.22 | C | 0.1 | 0.7 | 0.12 | C | 0.41 | 1.62 |
| rs11536898 | −3284 | 0.19 | A | 0.13 | 0.76 | 0.33 | A | 0.13 | 0.81 | 0.35 | A | 0.14 | 0.56 |
| rs1554973 | −3886 | 0.31 | C | 0.29 | 0.87 | 0.16 | C | 0.25 | 0.8 | 0.82 | T | 0.38 | 1.08 |
| rs7044464 | −4471 | 0.32 | A | 0.15 | 0.84 | 0.49 | A | 0.15 | 0.89 | 0.45 | A | 0.15 | 0.64 |
| rs7856729 | −4930 | 0.62 | T | 0.16 | 0.93 | 0.47 | T | 0.15 | 0.88 | 0.89 | T | 0.23 | 1.04 |
| rs7846989 | −6014 | 0.5 | C | 0.12 | 0.87 | 0.22 | C | 0.09 | 0.71 | 0.73 | C | 0.33 | 1.12 |
| rs7860896 | −6177 | 0.44 | G | 0.13 | 0.86 | 0.22 | G | 0.09 | 0.71 | 0.93 | G | 0.38 | 1.03 |
| rs7037117 | −6737 | 0.27 | G | 0.31 | 0.86 | 0.14 | G | 0.26 | 0.79 | 0.78 | A | 0.35 | 1.1 |
| rs913930 | −7083 | 0.003 | G | 0.31 | 1.48 | 0.0006 | G | 0.36 | 1.62 | 0.62 | G | 0.17 | 0.78 |
| rs1330305 | −7233 | 0.04 | C | 0 | 2.35 | Monoallelic | T | 0 | 0.31 | C | 0.02 | 1.84 | |
| rs1927905 | −8382 | 0.92 | C | 0.07 | 0.98 | 0.19 | C | 0.06 | 0.67 | 0.2 | C | 0.17 | 1.44 |
| rs7045953 | −8869 | 0.57 | G | 0.17 | 0.92 | 0.47 | G | 0.15 | 0.88 | 0.99 | G | 0.26 | 1.01 |
| rs7020005 | −10141 | 0.83 | T | 0.01 | 0.89 | Monoallelic | C | 0 | 0.93 | T | 0.06 | 0.96 | |
| rs10759934 | −12070 | 0.03 | A | 0.47 | 0.75 | 0.03 | A | 0.49 | 0.75 | 0.56 | T | 0.19 | 0.79 |
- * Nucleotide numbers come from the ATG translational start site for 5′ SNPs and from the translational stop site for 3′ SNPs.
Subgroup analyses were also conducted by donor race. SNP rs913930 was associated with an increased hazard of LGF in Caucasian liver donors (HR = 1.62, P = 0.0006) but not in African American donors (HR = 0.78, P = 0.62). A haplotype block analysis showed that the region between rs913930 and the coding sequences of TLR4 exhibited increased recombination in African Americans and thus would have lower linkage disequilibrium (LD) between SNPs in African Americans versus Caucasians. A Kaplan-Meier curve showing the time to LGF by the rs913930 genotype is shown in Fig. 1. Graft survival among recipients with livers from Caucasian donors was significantly reduced with the TT donor genotype versus the CC genotype (HR = 2.66, 95% confidence interval = 1.59-4.45, P = 0.0002). None of the SNPs were significant in recipients of livers from African American donors, although 2 variants (rs11536865 and rs5030717) had suggestive P values (P = 0.002 for both SNPs) when multiple testing was taken into consideration. It should be noted that the number of genotyped African American liver donors was much smaller than the number of Caucasian liver donors (128 versus 610). An analysis of the SNPs with a multi-SNP model using the top 3 SNPs (rs913930, rs11536865, and rs5030717) showed that the association became less significant and revealed that there was no strong haplotype effect.
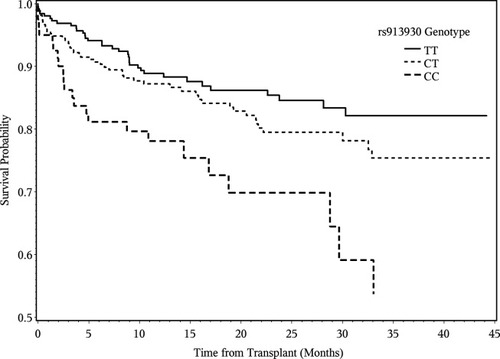
Time to all-cause LGF. Kaplan-Meier curves show the effects of rs913930 genotypes on graft survival in Caucasians only. Six hundred ten recipients received livers from Caucasian donors, and 112 (18%) experienced LGF.
A multivariate Cox proportional hazards model for LGF was also created (Table 3). The model was adjusted for the following: recipient race, donor race (Caucasian and African American donors only), recipient HCV status, first liver transplant, previous abdominal surgery, recipient BMI (linear and quadratic), and liver donor risk index (linear). The liver donor risk index had the most significant effect on LGF (P = 0.0003) and was previously associated with LGF.22 Other variables of modest significance were recipient HCV status (P = 0.03), first liver transplant (P = 0.036), and recipient BMI (quadratic; P = 0.0018).
| Variable | Group | Reference Group | HR (95% Confidence Interval) | P Value | Type 3 P Value |
|---|---|---|---|---|---|
| Donor race | African American | Caucasian | 0.95 (0.58-1.55) | 0.83 | 0.83 |
| Recipient race | African American | Caucasian | 1.21 (0.65-2.23) | 0.55 | 0.41 |
| Recipient race | Other | Caucasian | 1.36 (0.85-2.19) | 0.20 | |
| Recipient HCV status | Positive | Negative | 1.53 (1.04-2.24) | 0.032 | 0.03 |
| First liver transplant | No | Yes | 1.92 (1.04-3.53) | 0.036 | 0.04 |
| Previous abdominal surgery | Yes | No | 1.50 (1.01-2.22) | 0.044 | 0.04 |
| Recipient BMI (linear) | 1.02 (0.98-1.08) | 0.31 | 0.31 | ||
| Recipient BMI (quadratic) | 0.99 (0.99-1.00) | 0.0018 | 0.0018 | ||
| Liver donor risk index (linear) | 2.56 (1.54-4.26) | 0.0003 | 0.0003 |
We also included SNPs in our multivariate Cox proportional hazards model for LGF. None of the SNPs remained significant after we accounted for multiple comparisons with Bonferroni correction. However, SNPs rs11536865 (P = 0.00137) and rs5030717 (P = 0.00282) showed suggestive evidence for an association with LGF. We obtained similar results after the removal of pediatric recipients (n = 23).
DISCUSSION
We identified 3 SNPs within the deceased donor TLR4 gene that were significantly associated with LGF after multiple comparisons were taken into account: rs11536865 and rs5030717 in all donors and rs913930 only in Caucasian donors. For all 3 SNPs, the risk for graft loss increased when the minor allele was present. A Kaplan-Meier plot (Fig. 1) shows that for rs913930 in Caucasian donors, the risk increased with an increasing number of minor alleles. It is notable that SNP rs913930, located in the 3′ end of TLR4, was significant in Caucasians but not in African American donors. It is likely that rs913930 is not functional but is in LD with a functional SNP closer to or within the TLR4 gene. A haplotype block analysis showed that the region between this SNP and the TLR4 gene had a greater level of recombination in African Americans versus Caucasians. In the Caucasian liver donors, LD between rs913930 and the putative functional SNP was maintained, but LD was likely abolished in the African American donors because of more frequent recombination. Two previously reported functional variants, SNPs rs4986790 (p.Asp299Gly) and rs4986791 (p.Thr399Ile), were not associated with LGF (P = 0.43 and P = 0.8, respectively) in our data. It may be that their contribution to graft failure is minimal or is not associated with the donor genotype, our study is underpowered, or the reported associations were false positives.
Those SNPs identified in this study were associated with an increase in LGF. Possible mechanisms for an increased risk of graft loss include a variant that either increases the expression of TLR4 or increases messenger RNA stability and results in a higher level of TLR4 signaling of inflammation and thus organ damage and eventual loss.
The SNP rs10759930 was modestly associated with HCV infection (P = 0.052) in Caucasian donors, but the importance of this is unknown. It has been previously shown that the HCV status does not alter hepatic expression levels of TLR4,13 but the p.Arg753Gln polymorphism has been associated with allograft failure after liver transplantation for chronic HCV.23
Our study has several limitations. The SRTR does not provide detailed information about the causes of LGF, so it is not possible to determine whether infectious complications were involved in the development of LGF. The SRTR also does not provide information about the recurrence of HCV after liver transplantation. Therefore, it is not possible to determine the role of TLR4 SNPs in the occurrence of infectious complications or in the recurrence of HCV.
Much work has been done on the identification of recipient genetic variants affecting allograft outcomes, but genetic variation in the donor genome will also most likely affect the outcomes of grafts. Donor polymorphisms in TLR4 could act by modulating TLR4 activity and, therefore, affect the risk of graft loss. Additionally, there is a suggestion for an interaction between TRL4 SNPs and HCV status.
Acknowledgements
The authors thank the families who agreed to participate in this study. They also thank the following organ procurement organizations that provided donor samples: LifeSource (St. Paul, MN), LifeQuest (Gainesville, FL), New Jersey Organ & Tissue Sharing Network (New Providence, NJ), Organ Donor Center of Hawaii (Honolulu, HI), Southwest Transplant Alliance (Dallas, TX), One Legacy (Los Angeles, CA), New England Organ Bank (Waltham, MA), Lifebanc (Cleveland, OH), and Louisiana Organ Procurement Agency (Metairie, LA).