Substrate quality affects microbial- and enzyme activities in rooted soil†
Focus Issue “Soil processes regulation at hot ‘spots'”. Selected papers presented on a workshop organized by the German Soil Science Society (DBG), Commissions II, III, and VII held in Freising-Weihenstephan (Germany), May 4–6, 2014 (Conveners: Carsten W. Müller, Thilo Eickhorst, Markus Steffens, and Sven Marhan).
Abstract
The rhizosphere reflects a sphere of high substrate input by means of rhizodeposits. Active microorganisms and extracellular enzymes are known to be responsible for substrate utilization in soil, especially in rooted soil. We tested for microbial- and enzyme activities in arable soil, in order to investigate the effects of continuous input of easily available organics (e.g., root-exudates) to the microbial community. In a field experiment with maize, rooted and root-free soil were analyzed and rhizosphere processes were linked to microbial activity indicators such as specific microbial growth rates and kinetics of six hydrolytic extracellular enzymes: β-glucosidase, β-cellobiohydrolase, β-xylosidase, acid phosphatase, leucine- and tyrosine-aminopeptidase.
Higher potential activities of leucine-aminopeptidase (2-fold) for rooted vs. root-free soil suggested increased costs of enzyme production, which retarded the specific microbial growth rates. Total microbial biomass determined by the substrate-induced respiration technique and dsDNA extraction method was 23% and 42% higher in the rooted surface-layer (0–10 cm) compared to the root-free soil, respectively. For the rooted soil, potential enzyme activities of β-glucosidase were reduced by 23% and acid phosphatase by 25%, and increased by 300% for β-cellobiohydrolase at 10–20 cm depth compared to the surface-layer. The actively growing microbial biomass increased by the 17-fold in rooted soil in the 10–20 cm layer compared to the upper 10 cm. Despite the specific microbial growth rates showing no changes in the presence of roots, these rates decreased by 42% at 10–20 cm depth compared to the surface-layer. This suggests the dominance in abundances of highly active but slower growing microbes with depth, reflecting also their slower turnover. Shifts in microbial growth strategy, upregulation of enzyme production and increased microbial respiration indicate strong root effects in maize planted soil.
1 Introduction
The rhizosphere is considered as one of the most important microbial hotspots, more precisely a ‘hot sphere' in soil, as it is characterized by high microbial abundance and activity due to high amounts and diversity of easily available substrates (Hinsinger et al., 2005; Walker et al., 2003). There are three main sources of substrate input to the rhizosphere: (1) root exudates released from intact cells, (2) lysates of sloughed-off cells and root tissue, and (3) mucilage (Gregory, 2006; Neumann and Römheld, 2007). These forms of root-derived C are frequently termed rhizodeposition. Root exudates are readily available sources of C and energy for microbes (Haichar et al., 2008; Paterson, 2003; Paterson et al., 2007).
The release of labile compounds (including enzymes) by living roots or by lysis of root cells stimulates microbial activity (Nannipieri et al., 2012) and microbial growth (Panikov, 1995; Oger et al., 2004; Blagodatskaya et al., 2009) in the similar ways as rhizodeposits (Kuzyakov and Domanski, 2000; Marschner et al., 2004). The release of root exudates and other rhizodeposits is ongoing and is localized in soil (Pausch and Kuzyakov, 2011). Consequently, localization of easily available C produces hotspots of microbial abundance and activities, frequently termed as the “rhizosphere effect” (Lynch, 1990; Sørensen, 1997).
It is thought that the production of extracellular enzymes is regulated by nutrient availability and energy demand (Sinsabaugh et al., 2009). Therefore, extracellular enzyme activities in the rhizosphere are generally higher compared to root-free soils, similarly to total microbial biomass and microbial activity measured as respiration or growth rates (Badalucco and Nannipieri, 2005). Roots and associated mycorrhizal community are known to be major producers of β-glucosidases and acid phosphatases (Conn and Dighton, 2000). Despite soil enzymes being partly of plant origin, the microorganisms are the main source of enzymes mediating the cycling of major nutrients (C, N, P, and S) (Aon et al., 2001) and, thus, enzyme activity is frequently proportional to microbial biomass (Frankenberger and Dick, 1983). Hence, overall greater microbial biomass and higher enzyme activity can be predicted not solely in the rhizosphere but in a whole soil layer with high root density, e.g., in rooted soil as compared with soil without plants, e.g., in bare fallow soil.
The upper 30 cm contain 70–90% of the root biomass of maize (Amos and Walters, 2006), where available C sources induce activity of numerous microbial groups which are usually limited by N. Nutrient limitation for roots and microorganisms in the rhizosphere is far greater than in root-free soil. This leads to strong competition between roots and microorganisms for nutrients (Paterson, 2003; Kuzyakov and Xu, 2013). Hence, the rhizosphere is not only a hotspot of microbial activity, but also a hotspot of plant-microbial interactions including competition, resulting not only in acceleration but under specific conditions also in retardation of microbial growth (Blagodatskaya et al., 2014b).
As microbial communities allocate resources to enzyme production in relation to substrate availability and growth requirements to reduce costs and maximize their resource returns (Allison and Vitousek, 2005), we hypothesized that specific microbial growth rates increase in rooted soil compared to the fallow control. We suggested that plant-induced lower inorganic N contents in the soil compared to fallow control increase activities of peptidases (Stursova et al., 2006). We further hypothesized that enzyme activity per unit microbial biomass (e.g., specific activity) would increase from 0–10 cm to 10–20 cm, reflecting greater microbial allocation to C-cycling enzyme production depending on decreased C availability (Allison et al., 2011).
These hypotheses were tested in a multi-factorial field manipulation experiment with soil sampled under maize (rooted soil) and bare fallow at two depths (0–10 cm and 10–20 cm). Potential enzyme activities and soil microbial biomass were measured. Microbial growth was determined by a kinetic approach due to the substrate-induced respiratory response of microorganisms, enabling estimation of total and growing biomass of the glucose-consuming part of microbial community (Panikov, 1995; Panikov and Sizova, 1996). We used substrate-induced respiration (SIR) (Anderson and Domsch, 1978) and substrate-induced growth respiration (SIGR) of microbial cells. By combining these methods we were able to investigate microbial activity in the rhizosphere in order to elucidate the effects of rhizodeposits on microbial activity.
2 Material and methods
2.1 Study site
The experimental agricultural field is located on the terrace plain of the river Leine in the NW of Göttingen (Lower-Saxony, Germany, 51°33´N, 9°53´E; 158 m asl). The area has a temperate climate with a long-term mean annual precipitation of 645 mm and a mean annual air temperature of 8.7°C. The dominant soil types are Luvisols.
In spring 2012, 12 experimental field plots (5 m x 5 m) were established and separated from each other by buffer stripes of 2 m and 6 m in row and inter-row, respectively. Two treatments, rooted (P) and root-free (F) soil, were set up on the experimental plots with 4 replicates each. For rooted soil, hybrid maize (Zea mays L., Codisco/TMTD 98% Satec) was sown on 4 plots at a density of twelve plants per square meter in April 2013. In addition 4 plots remained unplanted as a bare fallow control. The fallow control plots were shaded with blinds (mechanical shading 50% and 80%; Accura NTV oHG, Heidenheim). To accomplish similar environmental conditions between the plots, the shading level represented a mean leaf area index of plants during the vegetation period.
2.2 Sampling and preparation
In July 2013, we sampled the soil at two depths (0–10 cm, 10–20 cm) for each plot. The field moist soil samples were frozen at −18°C until analysis. Freezing is known to influence the enzyme activities of extracellular hydrolytic enzymes (Gianfreda and Ruggiero, 2006; Lee et al., 2007). Following the study of German et al. (2011), we considered, however, that freezing would not affect the comparability of rooted vs. root-free soil as all soil samples were frozen and treated similarly.
Prior to analysis, soil samples were thawed in the refrigerator, sieved (< 2 mm), and fine roots and other plant debris were carefully removed with tweezers. The sieved field-moist soil samples were pre-incubated for 72 h at 22°C. Soil sub-samples of each plot and depth were dried at 105°C (24 h) to determine total C (Ct), total N (Nt), and moisture content. The moisture contents of the soil samples ranged from 14% for rooted to 18% for fallow soil. Prior analysis the moisture content was adjusted to 60% of the water holding capacity (WHC). No significant differences were detected in pH, Ct, or Nt content of rooted and root-free sampled soil.
The measurements of microbial respiration, such as SIR and SIGR, were used to determine microbial biomass and active microbial biomass as well as microbial growth rates in rooted vs. root-free soil to exhibit the responses of microbes to root exudation. Additionally, we determined the dsDNA-extracted microbial biomass C for validation. The potential hydrolytic extracellular enzyme activities were determined in order to elucidate enzyme production strategies of microorganisms due to substrate decomposition.
2.3 dsDNA extraction and quantification procedure
Total soil DNA was extracted by the FastDNA® SPIN kit for soil (MP Biomedicals, Germany). Extraction procedure was carried out according to the manufacturer's protocol with 0.5 g of pre-incubated soil. The method of DNA isolation involved bead beating procedure and binding of DNA to the silica matrix. Before extraction, soils were placed into a freezer overnight to ensure higher DNA yields. Thereafter, soils were added to lysing tubes, treated with lysis buffer, subjected to bead beating in the FastPrep® instrument, and processed by protein precipitation solution. DNA was bound to a silica matrix, washed, and eluted in DNase-free water.
Cmic (µg g−1 soil) = FDNA × dsDNA (µg g−1 soil).
(1)2.4 Enzyme assays
By the use of 4-methylumbelliferone-β-D-cellobioside, 4-methylumbelliferone-β-D-glucoside, 4-methylumbelliferone-phosphate, 4-methylumbelliferone-7-β-D-xyloside, L-leucine-7amino-4-methylcoumarin hydrochloride, and L-tyrosine-7amino-4-methylcoumarin, the enzyme activities of β-cellobiohydrolase (exo-1,4-β-glucanase, EC 3.2.1.91), β-glucosidase (EC 3.2.1.21), acid phosphatase (EC 3.1.3.2), β-xylosidase (EC 3.2.2.27), and leucine-/tyrosine-aminopeptidase (EC 3.4.11.1) were determined, respectively. Half a gram field moist soil was added to 50 mL sterile water in autoclaved jars. Aliquots of 50 µL were withdrawn and dispensed in 96-well microplates (Brand pureGrade, black) while stirring the suspension. Buffer (80 mL) was added (0.1 M MES buffer, pH 6.1 for carbohydrases and phosphatase, 0.05 M TRIZMA buffer, pH 7.8 for leucine-/tyrosine-aminopeptidase) (Marx et al., 2005). Finally, 100 µL of series concentrations of substrate solutions (20, 40, 60, 80, 100, 200, 400 µmol substrate g soil−1) were added to the wells. Plates were kept at 21°C, agitated, and measured fluorometrically (excitation: 360 nm; emission: 450 nm) after 1 h, 2 h, and 3 h incubation with an automated fluorometric plate-reader (Wallac 1420, Perkin Elmer, Turku, Finland). Fluorescence was converted into an amount of MUB (4-methylumbelliferone) or AMC (7-amino-4-methylcoumarin), according to specific standards, which had been prepared in sub-samples from the various soil suspensions. The kinetic parameter, Vmax, was estimated using non-linear regression techniques (Michaelis-Menten kinetics) (Marx et al., 2005). Each field replicate was measured as an analytical triplicate.
2.5 Substrate-induced growth respiration and calculation of growth parameters
The substrate induced growth respiration (SIGR) method was conducted in a climate chamber (16°C). Therefore, 23 g of each pre-incubated and moistened (WHC 60%) soil sample was incubated in a microcosm after addition of the substrates and nutrients (Blagodatsky et al., 2000). The amended substrate mixture contained glucose (10 mg g−1) and mineral salts, e.g., 1.9 mg g−1 (NH4)SO4, 2.25 mg g−1 K2HPO4, and 3.8 mg g−1 MgSO4 · 7 H2O. Instead of talcum, a glucose solution was applied. Glucose was used, because it is one of the abundant components of root exudates (Whipps and Lynch, 1983; Derrien et al., 2004). Substrate concentrations, sufficient for unlimited exponential growth of microorganisms, were estimated in preliminary experiments in which different amounts of glucose and nutrients were added. The amount of mineral salts was selected so that the added substrate did not change the pH of soil (< 0.1) (Blagodatskaya et al., 2007). After addition of the substrate-nutrient mixture and stirring with a common, handheld kitchen blender, the soil samples were immediately placed into 24 flasks (394 cm3) (Anderson and Domsch, 1978). A gas chromatograph (GC 6000 VEGA series 2, Carlo Erba instruments, UK) was modified for automatic sampling, measuring and calibration. The soil samples were kept in closed systems (microcosm) under quasi-stationary conditions and the evolved CO2 was measured every 120 min.
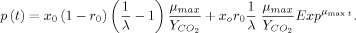 (2)
(2)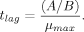 (3)
(3)The following unlimited exponential growth phase is dominated by a growing, active microbial biomass. More complete theoretical background and details on equations derivations were described elsewhere (Blagodatsky et al., 2000; Panikov, 1995; Wutzler et al., 2012).
2.6 Substrate-induced respiration
The substrate-induced microbial respiration (SIR) method provides a parameter for the potentially active microbial biomass without any growth of microbial cells based on respiration measurements following the addition of glucose and mineral salts as it is already explained for SIGR. The same amount of soil was incubated in flasks (1098 cm3) for 5 h after addition of the substrates. Gas samples (15 mL) were taken hourly and the C concentrations were analyzed by gas chromatography (GC 6000 VEGA series 2, Carlo Erba instruments, UK). We obtained the CO2 concentrations and calculated the CO2 flux rates. The data were corrected by the specific gas flux factor and multiplied with the headspace volume. Afterwards, the CO2 fluxes were related to the dry weight of the soil and time during the incubation experiment (Anderson and Domsch, 1978).
2.7 Salt-extractable and total N
Moist soil (7.5 g) was extracted with 30 mL of 0.05 M K2SO4 for 1 h (Bruulsema and Duxbury, 1996) by overhead shaking (40 rev min−1). The soil suspension was centrifuged for 10 min at approx. 2,500 g. Afterwards, the supernatant was filtered through Rotilabo-rondfilters (type 15A, Carl Roth GmbH & Co. KG, Karlsruhe, Germany). The N concentrations in the K2SO4 extracts were measured using a multi N/C analyzer (multi N/C analyzer 2100S, Analytik Jena, Germany). The total N-contents were measured using a elemental analyzer (NA1110, CE –instruments, Rodana, Milano, Italy).
2.8 Statistical analyses
The means of four field replicates with standard errors are presented in tables and figures. A t-test was applied to characterize the effects of roots and soil depths. When significant effects were identified, a multiple post-hoc comparison using the Holm-Sidak method (P < 0.05) was performed.
Parameter optimization was restricted to the applied model Equation (2) as indicated by maximum values of statistic criteria: r2, the fraction of total variation explained by the model defined as ratio of model weighted sum of squares to total weighted sum of squares. Outliers were identified by the ROUT method, based on the False Discovery Rate (FDR), where Q got specified, which was the maximum desired FDR (Motulsky and Brown, 2006). The data of potential enzyme activities was treated in the same way.
3 Results
3.1 The rhizosphere effect
Microbial biomass C (dsDNA derived) in the surface-layer was 42% higher in rooted vs. root-free soil. This was confirmed by higher activities of β-glucosidase (4.7-fold) and leucine-aminopeptidase (2-fold) as well as by SIR (23%) in the rooted surface-layer compared to root-free soil. Higher specific enzyme activities (potential enzyme activity per DNA content) were observed for leucine-aminopeptidase and β-glucosidase in rooted compared to root-free soil (Table 2). Microbial biomass C based on the DNA content [Eq. 1, Fig. 1] showed the same trends as that assessed by SIR for rooted and root-free soil in the uppermost 10 cm depth [Eq. 4, Fig. 2]. No significant differences were detected for the maximum specific growth rates (µmax) and β-xylosidase between rooted and root-free soils. Total N- and salt-extractable N-contents reduced for rooted soil compared the fallow control (Fig. 5). Especially the K2SO4-extractable N-contents decreased by 26% in the first 10 cm and 53% at 10–20 cm depth for rooted vs. root-free soil.

Extractable dsDNA contents (DNA, ± SEM) in root-free (F) and rooted (P) soil at two depths (0–10 cm, 10–20 cm). The dsDNA-Cmic contents were calculated using a factor of 5.02 (Anderson and Martens, 2013). Significant root effects are indicated by different capital letters (P < 0.05).
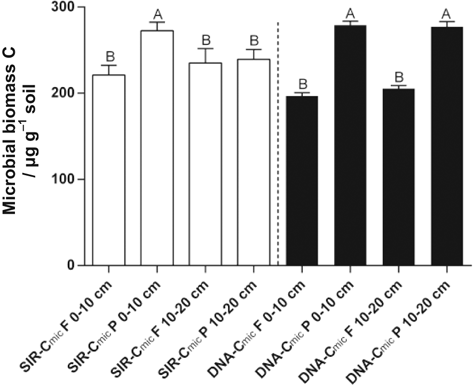
Comparison of substrate-induced respiration (SIR)-Cmic contents and dsDNA-Cmic contents (Cmic, ± SEM) for root-free (F) and rooted (P) soil at two depths. Significant root effects comparing SIR-Cmic contents and DNA-Cmic contents are indicated by different capital letters (P < 0.05).
3.2 Effects on microbial indicators with soil depth
The effect of depth on microbial parameters was more pronounced in rooted vs. root-free soil. Microbial biomass C decreased by 14% with depth in rooted soil (Table 1; SIR derived). The β-glucosidase activity was reduced by 23% and acid phosphatase by 25% for the rooted soil at 10–20 cm depth compared to the surface-layer (Fig. 3). The β-cellobiohydrolase activities in rooted soil almost tripled for rooted soil at 10–20 cm compared to 0–10 cm depth. No clear pattern with depth for C-cycling specific enzymes was determined (Table 2). Despite the microbial specific growth rates were independent on the root presence, these rates significantly slowed down for 42% in the lower layer compared to the first 10 cm depth (Fig. 4).
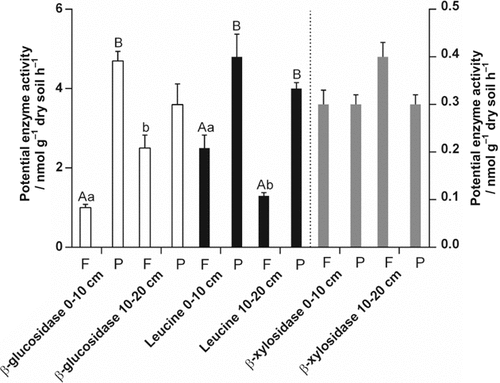
Potential hydrolytic exo-enzyme activities (Vmax; ± SEM, n = 12) for root-free (F) and rooted (P) soils at two depths (0–10 cm, 10–20 cm) are presented. Significant root effects are indicated by different capital letters. Lower-case letters signed significant root effects with depth (P < 0.05).
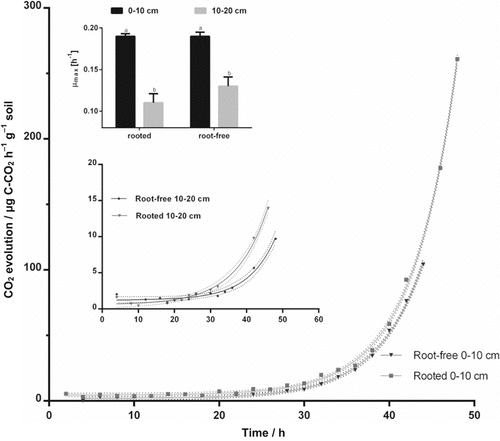
Maximal specific growth rates (µmax, ± SEM) are presented for rooted and root-free soil at depth intervals 0–10 cm and 10–20 cm. Significant effects are assessed by Mann–Whitney test (P < 0.05) and indicated by different capital letters. Lower-case letters signed significant root effects with depth (P < 0.05). A confidence band (95%) was calculated for rooted and root-free soil to be aware of outlier.
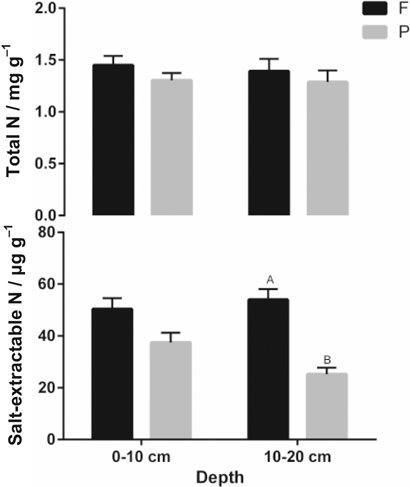
Total N and salt-extractable N (N, ± SEM) are presented for rooted (P) and root-free soil (F) at depth intervals 0–10 cm and 10–20 cm soil depth. Significant root effects are indicated by different capital letters (P < 0.05).
| Soil | SIRa | Active growing Cmic | Lag time | Total cell mass | |
|---|---|---|---|---|---|
| / µg CO2-C g−1 h−1 | / µg C g−1 | / % | / h | / µg g−1 | |
| Rooted 0–10 cm | 9.1 ± 0.2 | 0.2 ± 0.01a | 0.1 | 27 | 619 ± 10 |
| Root-free 0–10 cm | 7.4 ± 0.5 | 0.3 ± 0.03 | 0.2 | 23 | 437 ± 9 |
| Rooted 10–20 cm | 8.0 ± 0.2 | 3.5 ± 0.3Ab | 1.3 | 18 | 616 ± 12 |
| Root-free 10–20 cm | 7.8 ± 0.3 | 0.6 ± 0.07B | 0.3 | 30 | 456 ± 8 |
- aSIR = Substrate-induced respiration.
| Soil | Potential enzyme activity to dsDNA ratios / nmol h−1 mg−1 dsDNA | |||||
|---|---|---|---|---|---|---|
| Tyrosine | Acid phosphate | β-xylosidase | Leucine | β-cellobiohydrolase | β-glucosidase | |
| Root-free 0–10 cm | 23.0 ± 0.4a | 107.1 ± 1.2aA | 7.7 ± 0.1 | 64.8 ± 1.6aA | 132.6 ± 2.5aA | 25.5 ± 0.4aA |
| Rooted 0–10 cm | 18.0 ± 0.3a | 64.8 ± 0.9aB | 5.4 ± 0.1 | 86.4 ± 1.9aB | 16.2 ± 0.1aB | 84.6 ± 0.9aB |
| Root-free 10–20 cm | 31.8 ± 0.5b | 85.6 ± 1.3bA | 9.8 ± 0.1 | 31.8 ± 0.4bA | 58.7 ± 1.9bA | 61.1 ± 1.5b |
| Rooted 10–20 cm | 29.0 ± 0.3b | 48.9 ± 0.6bB | 5.4 ± 0.1 | 72.5 ± 0.7bB | 48.9 ± 1.5bB | 65.2 ± 1.8b |
The maximum specific microbial growth rates (µmax) varied between 0.11 ± 0.015 and 0.19 ± 0.03 h−1 (Eq. 2) overall the soil samples (Fig. 4). The actively growing microbial biomass did not exceed 1.3% of total biomass and was highest in rooted soil at 10–20 cm depth. Active part doubled with depth for root-free and increased by 17.5 times for rooted soil with the depth. In rooted soil microorganisms started to grow 12 h earlier at 10–20 cm depth compared with root-free soil (Table 1).
4 Discussion
4.1 Microbial biomass, growth, and activity in the rhizosphere and root-free soil
The abundance of roots clearly enhanced microbial biomass by increased rhizodeposition. A portion of 30–60% of the photosynthetically fixed C can be translocated to the roots and up to 40% of the fixed C can be lost by rhizodeposition (Kuzyakov and Domanski, 2000; Neumann and Römheld, 2007). At our field site about 50% of the roots were allocated to the upper 10 cm (Pausch et al., 2013). The decreasing root biomass with depth led to lower rhizodeposition (Pausch et al., 2013), which reflected a positive correlation (Van der Krift et al., 2001). As a consequence of lower root biomass and rhizodeposits, microbial turnover increased and specific growth rates retarded at 10–20 cm depth compared to the surface-layer (Blagodatskaya et al., 2014b).
The decreased input of easily decomposable substrates by rhizodeposition with depth may induce stronger competition for substrates between microorganisms, especially for N (Fontaine et al., 2003; Paterson, 2003; Fischer et al., 2013), which resulted in lower microbial biomass at 10–20 cm depth (Badalucco and Nannipieri, 2005) or in slower growth (Merckx et al., 1987). Slower growth rates but greater fraction of active biomass and higher activity of cellulases at 10–20 cm depth vs. the first 10 cm depth indicated the shift in abundances to slow-growing oligotrophic microorganisms (Blagodatskaya et al., 2014a, 2014c). This was related to the high abundance of cellulolytic enzymes, which was possibly associated with dead roots. The decrease of microbial specific growth rates with the depth could be a consequence of growth limitation by the depletion of N (Helal and Sauerbeck, 1986; Merckx et al., 1987). The competition for N between microbe-microbe or plant-microbe interactions in rooted soil (Bowen and Rovira, 1999; Paterson, 2003; Badri et al., 2009) may have shifted the microbial community structure from r-selected to slow-growing (K-selected) microorganisms (Blagodatskaya et al., 2009; Fierer et al., 2007).
The 23% greater microbial respiration response in rooted vs. root-free soils indicated a high fraction of potentially active microorganisms (Blagodatskaya and Kuzyakov, 2013). Microbial cells, which maintain a potentially activity status (Van Bodegom, 2007), are able to immediately utilize an occasional substrate input (De Nobili et al., 2001). This was confirmed by a shorter lag-period for rooted soil at 10–20 cm depth, reflecting a high microbial affinity to the respective substrate input.
4.2 Potential and specific enzyme activities in rooted soil
The increased activities of leucine-aminopeptidase for rooted soil compared to the fallow control indicated the higher energy investments of microbes in producing proteolytic exo-enzymes (e.g., leucine-aminopeptidase) in order to utilize N-bonded molecules. This suggests that lower contents of inorganic N are available for microbes in rooted soil (Stursova et al., 2006). Conformingly, the salt-extractable N- and total N-contents were reduced in the rooted sphere, especially at 10–20 cm depth. Due to reduced accessibility to proteolytic degradation, immobilized enzymes often demonstrate higher stability compared with free extracellular enzymes (Allison, 2006). Plant roots as sinks for excess N enhanced continued mineralization driven by microbes but shifted interaction of enzymatic systems (Pinton et al., 2007).
As the increased costs for enzyme production reduce the fitness of microbes, because those resources cannot be invested for reproduction (Allison et al., 2011), the specific microbial growth rates were retarded in rooted vs. root-free soil (Blagodatskaya et al., 2014b).
The leucine-aminopeptidase and β-glucosidase activities decreased for rooted soil at the first 20 cm depth, which was in line with the studies of Steinweg et al. (2013) and Taylor et al. (2002). However, the β-cellobiohydrolase activity increased and β-xylosidase activity stayed constant in rooted soil at the first 20 cm depth. Enzymatic systems of β-cellobiohydrolases and β-glucosidases showed a contra-balanced behavior, especially in the surface-layer. For substrate utilization it is suggested that soil microorganisms use glucan (cellulose) as the preferred substrate irrespective of the type of residue (Leitner et al., 2012; Amin et al., 2014).
In 10–20 cm depth, the ratios of β-xylosidase and β-glucosidase to dsDNA contents raised for root-free soil indicated lower availability of C sources for enzymatic C utilization compared to the first 10 cm (Stone et al., 2014). This is in accordance with the specific activities of C-cycling enzymes, reported to increase with depth and reflecting greater microbial allocation to C-cycling enzyme production depending on decreased C availability (Allison et al., 2011).
5 Conclusions
The applied combination of the three approaches analysis of the double-stranded DNA contents, enzyme activities, and respiration kinetics gave quantitative insights in microbial traits in rooted vs. root-free soil. Strong rhizosphere effects were elucidated for most of the measured microbial activity indicators. Thus, rooted soil had greater microbial biomass, potential enzyme activity rates, and substrate-induced respiration compared to root-free soil. Similar specific microbial growth rates and dsDNA-derived microbial biomass contents were demonstrated for rooted vs. root-free soil when we compared the two soil layer. However, the active microbial biomass increased strongly in the rhizosphere at the 10–20 cm depth.
The demand for N by microbes and maize plants clearly affected the potential and specific enzyme activities in the rooted sphere of an arable soil. Thus, the competition for that resource induced strong microbial- and plant-interactions, which boosted proteolytic enzyme activities (e.g., leucine-aminopeptidases) and hampered microbial growth in rooted soil. We conclude that the rhizosphere, namely rooted soil, serves as an area for microbes and extracellular enzymes, which strongly depend on the substrates present in the rooted zone of maize plants.
Acknowledgements
We would like to thank Susann Enzmann, Heike Strutz, and Viola Schade for their intensive help during the field and laboratory work. This study was supported by the German Research Foundation (DFG) within the Research Unit FOR 918 “Carbon Flow in Belowground Food Webs Assessed by Isotope Tracers”. Furthermore, we thank for the organization of the DBG Workshop: “Soil Processes—Is the Whole System Regulated at Hot Spots?—From Micro-scales to the Pedon”.




