Changes in the Salmonella enterica Enteritidis phenotypes in presence of acyl homoserine lactone quorum sensing signals
Abstract
Quorum sensing is used by bacteria to coordinate gene expression in response to population density and involves the production, detection and response to extracellular signaling molecules known as autoinducers (AIs). Salmonella does not synthesize the AI-1, acyl homoserine lactone (AHL) common to gram-negative bacteria; however, it has a receptor for AI-1, the SdiA protein. The effect of SdiA in modulating phenotypes of Salmonella has not been elucidated. In this report, we provide evidence that the AIs-1 affect Salmonella enterica serovar Enteritidis behavior by enhancing the biofilm formation and expression of virulence genes under anaerobic conditions. Biofilm formation by Salmonella was detected by the crystal violet method and by scanning electron microscopy. The presence of AHLs, particularly C12-HSL, increased biofilm formation and promoted expression of biofilm formation genes (lpfA, fimF, fliF, glgC) and virulence genes (hilA, invA, invF). Our results demonstrated that AHLs produced by other organisms played an important role in virulence phenotypes of Salmonella Enteritidis.
Abbreviations
-
- AHL
-
- acyl homoserine lactones
-
- AIs
-
- autoinducers
-
- C12 HSL
-
- homoserine lactone
-
- LB
-
- Luria Bertani
-
- MM
-
- minimal medium salts
-
- SPI
-
- salmonella pathogenicity island
-
- TSB
-
- tryticase soy broth
-
- QS
-
- quorum sensing
Introduction
Much interest has been shown in the last two decades in the elucidation of the paradigm of communication between bacteria that are able to modify gene expression patterns in the response to a variety of extracellular signals. Intra and intercellular communication between bacterial populations, termed quorum sensing (QS), is studied as an important tool for the coordination of metabolic activities 1. The QS system involves the production, detection and response to small extracellular signaling molecules known as autoinducers (AIs) 2-4. It has been reported that a variety of bacterial phenotypes, such as bioluminescence, sporulation, biofilm formation, conjugation, motility, competence, antibiotic, and enzymes production are regulated in response to signaling molecules of QS systems 4-9. The potential role of QS in foodborne bacteria has been studied to elucidate the regulatory mechanisms of infectious processes and bacterial spoilage of food 5.
The presence of Salmonella in food is of major concern once this pathogen comprised the primary cause of foodborne diseases in many countries for over 100 years. Nontyphoidal Salmonella enterica serovars Typhimurium and Enteritidis are responsible for most Salmonella infections in Europe and America 10-13. This pathogen also leading the causes of hospitalization and death in humans annually in the United States 11 and, the estimated total cost associated with Salmonella illnesses could approach several billion dollars annually.
Salmonella has QS systems regulated by the following AIs: AI-1, AI-2, and AI-3 14. The system mediated by AI-1 is most frequently observed in intraspecific communications in gram-negative bacteria and involves N-acyl homoserine lactones (AHL) as the signal molecule 14. However, in Salmonella, as in Escherichia coli, the system mediated by AI-1 is incomplete because it only shows the response regulator protein SdiA 15. Michael et al. 16 confirmed that SdiA is used to detect only signals from other bacterial species because of the lack of the gene encoding the AHL synthase in the E. coli and Salmonella Typhimurium genomes. Phylogenetic analysis of nucleotides encoding the SdiA protein, and comparative alignment of its amino acids showed that the gene and protein are conserved in Salmonella but this pathogen also failures to produce the signal molecule AHL 17, 18. Genetic screening identify that four genes, srgA, srgB, srgC, and rck, in the pSLT plasmid encoding virulence genes in Salmonella Typhimurium were dependent on the expression of sdiA 15. The actual functions of srgA, srgB, and srgC have not been elucidated 16, but sequence analysis suggests that srgA encodes a homologous perispasmatic protein disulfide isomerase, DsbA, which is required for assembly of the P pilus; srgB, encodes a putative lipoprotein; srgC encodes a putative transcriptional deregulator type similar to AraC; and rck encodes a small protein of the outer membrane that is important for survival during infections in humans. The actual functions of srgA, srgB, and srgC have not been elucidated 16. However, Moretro et al. 18 showed that a SdiA mutant did not affect Salmonella virulence in infection models in mice, birds and cattle. Sperandio 19 observed that SdiA did not show beneficial activity for Salmonella Typhimurium during colonization in mice, however, when co-infection occurs with a pathogen producing AHL, for example, Yersinia enterocolitica, SdiA has functional activity.
Therefore, the aim of this study was to evaluate the role of AI-1 in biofilm formation and expression of virulence genes by Salmonella Enteritidis PT4 under anaerobic conditions. The results can contribute to clarify the virulence of the pathogen in conditions in which it intercepts and thereby decodes the message of others gram-negative competitors and develops strategies for survival and success.
Materials and methods
Bacteria and culture conditions
A strain of S. enterica serovar Enteritidis phagotype PT4 578, which was isolated from chicken breasts was used, courtesy of the Oswaldo Cruz Foundation – FIOCRUZ (Rio de Janeiro, Brazil). The QS mechanism of this isolate was characterized previous 17. The stock cultures were performed in Luria Bertani broth (LB) (tryptone 1%, yeast extract 0.5% and NaCl 0.4%), pH 7.4, containing 20% (v/v) sterile glycerol and were frozen at 20 °C. Before each experiment, the cells were cultured two consecutive times in LB broth and incubated at 37 °C for 18 h under anaerobic conditions. The effect of AHL in biofilm formation was performed to add these purified molecules to the culture medium at a final concentration of 50 nM. The AHLs were suspended in acetonitrile and added to the culture medium composed of 1% acetonitrile in accordance with Michael et al. 16. An identical volume of acetonitrile was added to the culture media as a control treatment.
Biofilm formation
The biofilm formation was initially assessed in polystyrene microplates of 96 wells in an anaerobic chamber Forma Anaerobic System (Thermo Fischer Scientific, Waltham, Massachusetts, USA) containing 90 % (v/v) N2, 5% (v/v) H2, and 5% (v/v) CO2. The incubation occurred at 37 °C for 24, 48, 72, and 96 h. During the incubation periods, the optical density of the cell culture was measured at 600 nm, and the crystal violet 0.1% (p/v), retained by the adhered cells after staining, was determined at 590 nm on a Thermoplate Reader (Molecular Devices, San Diego, California, USA) according to the method described by Conway et al. 20. The biofilm formation was evaluated under anaerobic conditions in minimal medium salts (MM) and Trypticase Soy Broth (TSB) in the presence of the following AIs added at concentration of 50 nM: N-hexanoyl homoserine lactone-C6-HSL (Fluka, Germany), N-octanoyl homoserine lactone-C8-HSL (Fluka), N-decanoyl homoserine lactone-C10-HSL (Sigma–Aldrich, Germany), and N-dodecanoyl homoserine lactone-C12-HSL (Fluka).
Additionally, the biofilm formation was evaluated on polystyrene coupons, with the dimensions of 1.0 × 1.0 × 0.1 cm, at 1.0 cm2 of the area, immersed in TSB contained a mixture of AHLs or only C12-HSL. The effect of furanones (Sigma–Aldrich, Germany), an antagonist of AHL, was also evaluated. A mixture of 3-methyl-2 (5H) furanone, 2-methyltetrahydro-3-furanone, 2 (5H)-furanone and 2,2-dimethyl-3(2H) furanone was prepared in acetonitrile to maintain a final concentration of 50 nM. The furanones were added to the media with or without 50 nM of C12-HSL.
Microscopy
The polystyrene coupons were removed from the culture media after 36 h, rinsed with a solution of 0.05 M potassium phosphate (pH 6.8–7.2) and fixed for 1 h at room temperature with 2.5% glutaraldehyde in 0.05 M sodium phosphate buffer. Subsequently, they were washed with 0.05 M sodium phosphate buffer 6 times for periods of 10 min each, followed by a dehydration step with 35, 50, 70, 80, 95, 100, 100, and 100% ethanol, each for 10 min. Then, the drying step was performed at the critical point in the Bal-Tec CPD 30, with metallization in a Sputter Coater SCD 010 (Balzers, Liechtenstein) and observation on a scanning electron microscope (LEO, model VP 1430, England).
Evaluation of gene expression
The culture of Salmonella Enteritidis PT4 was reactivated in LB broth incubated at 37 °C anaerobically, until a population of 105 CFU ml−1 was reached, and C12-HSL was added at final concentration of 50 nM. A control culture was added with the identical volume of acetonitrile. The culture was again incubated for 2 or 7 h. After the incubation, the culture was centrifuged to obtain the total biomass and to perform the subsequent extraction of the total RNA. RNA was extracted with Trizol® Reagent (Invitrogen, California, USA) according to the supplier's instructions. RNA concentrations were then standardized to 1 μg/µl in each sample prior to transformation into cDNAs. RNA quality was analyzed by electrophoresis on a 1% (w/v) agarose gel. The extraction efficiency was assessed by spectrophotometric quantification (Ultrospec 3000, Pharmacia Biotech, Buckinghamshire, England) based on the A260/A280 ratio and the concentration in ng /ml. The total RNA was purified for removal of the DNA. Starting at 1 μg of RNA, the synthesis of the first cDNA ribbon was performed according to the protocol recommended by the manufacturer. Before reverse transcription, RNAs were treated with DNAse. ADNase I (Sigma, St. Louis, MI, USA) treatment is performed to eliminate residual DNA (0.25 U/µl, 1 h, 37 °C). Deactivation of DNase I was achieved by heating at 70 °C for 15 min and total elimination of chromosomal DNA was checked by quantitative PCR. The Reverse Transcription step was performed by using the Improm II TM reverse Transcription System (Promega, USA), with random hexamer primer according to the manufacturer's instructions. Designed primers for quantitative PCR were 20–25 bases long, had a G/C content over 50% and a Tm of about 60 °C. The length of the PCR products ranged 100 bp (Table 1). The primer Express version 3.0 design software was used to select primer sequences. Quantitative real-time PCR assay was performed with SYBR GREEN I (Applied Biosystems, Foster City, CA, USA) in optical gratings in 96-well plates in a Bio-Rad C1000 Manager sequence detector system. The composition of the PCR mix for each sample was as follows: 1 µl of sample, 1 µl of each primers, 3 µl of Milli Q water and 6 µl of Sybr Green I Master Mix (Applied Biosystems). Amplification program is as follows: an initial denaturing step at 95 °C (10 min), followed by 40 cycles of 95 °C (15 s) and 60 °C (1 min). In each run a negative control was included. A melting curve was obtained from a first step starting from 50 to 95 °C, to control specificities of quantitative PCR reaction for each primer pair. Efficiency of amplifications was determined by running a standard curve with serial dilutions of cDNA. The efficiency (E) was calculated with the formula E = [10(1/−s)] × 100, where s is the slope of the standard curve. The normalized reporter value (Rn) represented measured fluorescence detected during a PCR cycle. The Ct (Critical Threshold) stood for the first PCR cycle during which the fluorescence value (Rn) was significantly distinct from the background and corresponded to a signal. For each experiment, the baseline above a signal was considered significant was n=0.1. Results were analyzed using the comparative critical threshold (ΔΔCt) method in which the amount of target RNA was adjusted to a reference (internal target RNA). The following equations were used (“calibrators” refers to reference conditions and “sample” defines addition AHL conditions): ΔCt = Ct of internal control – Ct of gene of interest; ΔΔCt = ΔCt of sample – ΔCt of calibrator; relative expression level = 2ΔΔCt.
| Primer name | Oligonucleotide sequences (5’–3’) |
|---|---|
| hilAF | GCTGCACCAGGAAAGCATTAA |
| hilAR | GCGAAGTCCGGGAATACATC |
| invAF | ACAGTGCTCGTTTACGACC |
| invAR | ACTGGTACTGATCGATAAT |
| invF | ATGATTAACGGCTAATTGGGTGA |
| invR | CGGAAAAGCGAAGAGTGAATTAC |
| glgCF | TGACCAGAACTGGCCTATCC |
| glgCR | CAGCGAGTTCAGCGTCATAC |
| fimFF | CCTAACACGTTACCGCTTCA |
| fimFR | CCGGCGTCATATTTTCTGA |
| fliFF | GAAGCCATTCTGTCGCCTAT |
| fliFR | TGTAGTGCTCTTCCGTCTGC |
| lpfAF | GATGCGACACTGGTTTCTGT |
| LpfAR | CGGTGTTCATTTCAACAAGC |
| gyrAF | CCAATACGTTCATGGCGTAAAG |
| gyrAR | GATTATGCGATGTCGGTCATTGT |
In order to determine an internal control, the fold changes were calculated using the method 2ΔΔCt as previously described 21.
Statistical analysis
The results obtained in the detection of biofilm formation by Salmonella Enteritidis PT4 in different culture media in the presence of different AI-1 was analyzed with an analysis of variance (ANOVA) followed by Tukey's test. Student's t test was used for the statistical analysis of the gene expression. Values less than 0.05 were considered statistically significant, and all the comparisons were performed using GraphPad Prism 5.00 (GraphPad Software, San Diego, California, USA) software.
Results
Biofilm formation by Salmonella Enteritidis PT4 in the presence of AI-1
During the 96 h period following the biofilm formation by Salmonella Enteritidis PT4 under anaerobic conditions, greater adhesion was observed when the culture was grown in TSB medium, which was nutritionally richer and more complex than the minimal salt medium (Fig. 1A–D). Adhesion was estimated by calculating the ratio of the absorbance of crystal violet extract and the optical density of the total cells. The addition of 50 nM of each of the AHLs from C6 to C12 had a significant stimulatory effect (p < 0.05) on the adhesion of the cells to the surface of polystyrene and, among the tested AHLs, C12 showed the highest induction of this phenotype at 72 h incubation (Fig. 1).
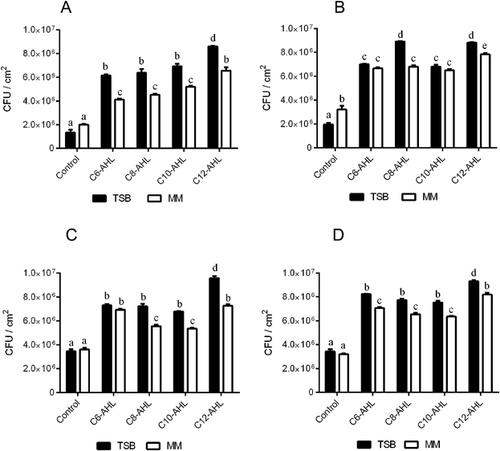
The effect of AI-1 on the cell adhesion of Salmonella Enteritidis PT4
Scanning electron microscopy allowed us to confirm the biofilm formation by Salmonella Enteritidis PT4 under the conditions studied, in the absence and presence of AI-1 (Fig. 2). TSB broth was selected to characterize the performance of AI on biofilm formation once it promoted increased cell adhesion. It was not possible to observe the typical biofilm structures on the surface of polystyrene coupons immersed in the Salmonella culture after 36 h of incubation in the culture medium to which AI-1 was not added (Fig. 2A). In this condition, it was possible to observe the presence of a few cells adhered. The adhesion of large numbers of cells on the coupons of polystyrene after 36 h of incubation was observed after a mixture of C6 to C12-HSL has been added to the culture medium, and it was possible to observe the presence of exopolymers (Fig. 2B). However, the presence of only C12-HSL resulted in additional aggregate of cells and the morphology characteristic of biofilm (Fig. 2C). On the observation of the image, it is possible to note multilayered cells that characterize bacterial biofilms (Fig. 2C). The compact biofilm structure in presence of C12-HSL was stable for 96 h (data not shown).
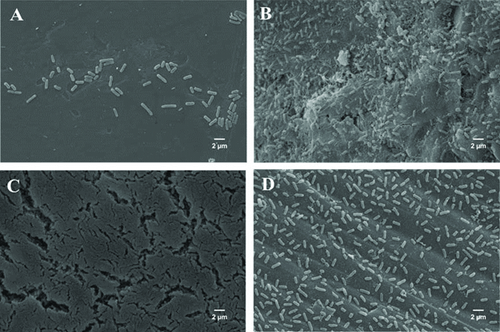
The presence of the furanones with C12-HSL resulted in adherent cells, with greater spacing between them; and a typical biofilm structure was not observed (Fig. 2D). Furanones are metabolites originally isolated from seaweed, Delisea pulchra, which are analogous to AHLs, with which they compete to bind to SdiA; however, they are unable to promote gene regulation. However, even in the presence of this QS inhibitor, a larger number of adhered cells (Fig. 2D) were observed when compared to the control (Fig. 2A).
Expression analysis by RT-qPCR of the genes involved in the biofilm formation
Figure 3 shows the relative expression of the glgC, fliF, lpfA, and fimF genes in Salmonella Enteritidis PT4 after incubation with C12-HSL for 2 h (Fig. 3A) and 7 h (Fig. 3B). The culture of Salmonella Enteritidis PT4 in absent of C12-HSL was used as the control and, the gyrA gene as the endogenous control. The dashed line in the graphs (Fig. 3) .shows the expression of the evaluated genes, equivalent to 1 in the cells of control. The addition of C12-HSL increased expression of the glgC gene, which is responsible for the regulation of glycogen biosynthesis proportionally to the increase of the incubation period (Fig. 3). Although the expression of the genes related to motor function, as fliF, and the fimbriae, as lpfA and fimF, was reduced after 2 h of contact with C12-HSL, there was a more significant increase (p < 0.05) and the expression was two times higher after 7 h of incubation in the presence of C12-HSL (Fig. 4). The addition of C12-HSL resulted in an increase in the expression (p < 0.05) of the glgC gene after 2 and 7 h of contact, with the longest incubation time resulting in an increase in the gene expression approximately 2 fold that of the control (p < 0.05) (Fig. 3).
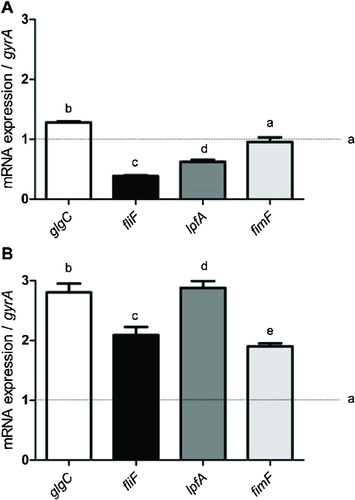
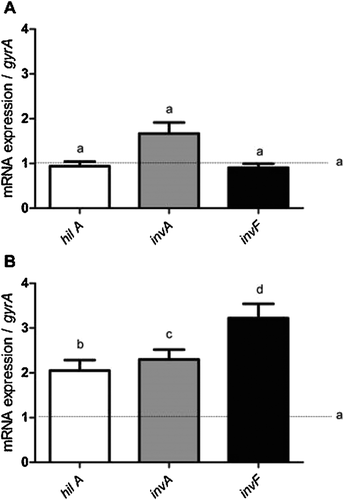
Analysis of the expression of the virulence genes by RT-qPCR
The expression of the virulence genes, hilA, invA, and invF, belonging to pathogenicity island 1 of Salmonella (SPI-1), was unchanged compared to that of the control at the end of the 2 h incubation in the presence of C12-HSL (Fig. 4A). Significant increases (p < 0.05) of up to 3.5 times in the expression of these genes were recorded in the presence of C12-HSL, with an increased incubation period of 7 h (Fig. 4B).
Discussion
The medium composition and environmental conditions affect the biofilm formation and this study showed that a complex medium induced greater adhesion of Salmonella Enteritidis PT4 in anaerobic condition. The results found in the literature are contradictory and, whereas some authors found that rich media increase the formation of bacterial biofilms 22, 23, others consider that the nutritional stress found in minimal media stimulate adhesion and subsequent biofilm formation by bacteria 22, 24, 25.
Although biofilm formation by many bacteria is related to the QS system mediated by AHLs, as in Pseudomonas aeruginosa 26, Burkholderia cepacia 27, Aeromonas hydrophila 28, Agrobacterium tumefaciens 29, and Hafnia alvei 25, in Salmonella this subject needs to be clarify. Indeed, the function of the receptor protein of AHLs in Salmonella remains unknown 30, 31. Dourou et al. 32 demonstrated that the growth of Salmonella Enteritidis may be affected by the presence of QS signaling compounds produced by other bacterial species. Liu et al. 33 found that AHLs increased rck expression in Salmonella carrying the pRST98 plasmid and suggested that probably this enhancing bacterial adherence, serum resistance and bacterial biofilm formation. Michael et al. 16 and Doulgeraki et al. 34 observed that the expression of sdiA can be changed depending on growth conditions, such as physical characteristic of the medium and temperature and, how this affect Salmonella behavior still remain to be understood.
Among the tested AHLs, C12-HSL promoted more compact and mature biofilm formation. The greater efficacy of the long carbon chain molecules in virulence phenotypes was reported by Rumbaugh et al. 35 in P. aeruginosa. Molecules with long carbon chain have been described particularly in communication between kingdoms, or eukaryotic and prokaryotic cells 36, 37.
Evidence of change in the characteristics of Salmonella Enteritidis PT4 in the presence of C12-HSL in anaerobic condition suggested the ability of this pathogen to survive and develop strategies to become more competitive. Considering the intestinal environment, with an abundant and diverse microbiota, Salmonella could perceive the presence of signals produced by competing bacteria and modify behavior, such as biofilm formation, to ensure their persistence in the environment 38.
One method to disrupt or manipulate the process of cell-cell signaling in bacteria by AI-1 is by the addition of similar molecules to AHLs, such as furanones. These substances could bind to the LuxR protein and their counterparts without activating them or even promote the degradation of these proteins and thus prevent the transcription of the genes involved in the regulation of biofilm formation 39, 40. In this study, the furanones inhibited the formation of mature biofilm without inhibiting adhesion. This result indicates that AHL probably does not regulate initial phase of biofilm formation by Salmonella and this observation was also done by Chorianapoulos et al. 41, who evidenced that several synthetic AHLs was not able to affect the early stages of biofilm formation.
The strong induction of the expression of the genes related to the biosynthesis of glycogen as glgC, which encodes an allosterically regulated enzyme that catalyzes the conversion of glucose-1-phosphate to ADP-glucose by the presence of C12-HSL, could promote the formation of biofilm. The regulation of glycogen biosynthesis in E. coli and Salmonella is highly interconnected with a wide variety of cellular processes and involves a complex set of factors that are adjusted to the physiological state of the cell, including the formation of biofilm 40.
The increased expression of the fliF, lpfA, and fimF genes in the presence of C12-HSL could be related as well to the increased capacity of biofilm formation. The fliF gene is present in the fliFGHIJK operon and encodes a protein of 61 kDa belonging to the basal body of the flagellum, the responsive motor function 42-44. The lpfA and fimF genes are part of the operons encoding the long type fimbriae and the type 1 fimbriae 45, respectively. These structures are important in pathogen adhesion to surfaces and the process in bacteria-host interactions in the colonization and invasion of intestinal cells for environmental persistence and biofilm formation 45.
The increased expression of genes in SPI-1 Salmonella Enteritidis PT4 after 7 h of incubation in the presence of C12-HSL suggested that AI-1 has a regulatory effect on the virulence of this pathogen. The hilABCD operon, present in SPI-1, encodes regulatory proteins, among them, HilA, which acts as a central regulator of the expression of genes related to invasion of the host cell by Salmonella 40, 46, 47.
The mechanism through which Salmonella spp. invades the host cell is quite complex and the bacteria promote its internalization by stimulation of phagocytic cells. One of the genes involved in this process is the invA gene located in the operon, invABCD, in SPI-1. This gene encodes a protein of periplasmic membrane of bacteria, InvA, and is essential for invasion of the host epithelial cells 48. The expression of genes of the SPI-1 and the next encoding effector proteins are translocated under the control of the regulatory protein, InvF, which is a member of the AraC/XylS family of transcriptional activators that, with a complex of the chaperone protein, SicA, regulate the expression of several genes 43, 49, 50. The invF gene is regulated by the HilA protein, which is encoded by a gene of the hil operon, SPI-1. HilA acts as a central regulator 43, 49.
In conclusion, this study provides strong evidence that the AHLs could stimulate biofilm formation and the expression of the SPI-1 genes of Salmonella. These results contribute to research that could provide additional information regarding the mechanisms of interspecies communication for future development of strategies to prevent biofilm formation by this pathogen. Knowledge of the regulation of virulence is an additional important approach because it could lead to procedures or therapies to reduce the pathogenic potential of these bacteria.
Acknowledgments
We thank the Center for Microscopy and Microanalysis, Federal University of Viçosa, and funding agency CNPq for financial the support.




