Contribution of HCN1 variant to sinus bradycardia: A case report
Funding information
This work has been supported by American Heart Association Grant-in-Aid (13GRNT16420018), National Institute of General Medical Sciences of the National Institutes of Health (U54GM104942), and a Research Development Grant from Health Sciences Center of West Virginia University to HY. The WVU Flow Cytometry & Single Cell Core Facility is supported by the WV IDeA grants P20GM121322 (CoBRE) and P20GM103434 (WV INBRE) and the S10 0D016265 (LSR Fortessa). Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number U54GM104942. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.”
Abstract
Background
Missense mutations in the hyperpolarization-activated cyclic nucleotide-modulated (HCN) channel 4 (HCN4) are one of the genetic causes of cardiac sinus bradycardia.
Objective
To investigate possible HCN4 channel mutation in a young patient with profound sinus bradycardia.
Methods
Direct sequencing of HCN4 and whole-exome sequencing were performed on DNA samples from the indexed patient (P), the patient's son (PS), and a family unrelated healthy long-distance running volunteer (V). Resting heart rate was 31 bpm for P, 67 bpm for PS, and 50 bpm for V. Immunoblots, flow cytometry, and immunocytofluorescence confocal imaging were used to study cellular distribution of channel variants. Patch-clamp electrophysiology was used to investigate the properties of mutant HCN1 channels.
Results
In P no missense mutations were found in the HCN4 gene; instead, we found two heterozygous variants in the HCN1 gene: deletion of an N-terminal glycine triplet (72GGG74, “N-del”) and a novel missense variant, P851A, in the C-terminal region. N-del variant was found before and shared by PS. These two variations were not found in V. Compared to wild type, N-del and P851A reduced cell surface expression and negatively shifted voltage-activation with slower activation kinetics.
Conclusion
Decreased channel activity HCN1 mutant channel makes it unable to contribute to early depolarization of sinus node action potential, thus likely a main cause of the profound sinus bradycardia in this patient.
1 INTRODUCTION
Disease of the sinus node and resultant impairment of atrioventricular conduction induce an excessively low ventricular rate (bradycardia), which cannot meet the needs of the organism.1 Sick sinus syndrome refers to intrinsic sinus node dysfunction that often results in symptomatic sinus bradycardia or sinus pauses.1 In certain states, profound bradycardia can be associated with ventricular arrhythmias leading to cardiac arrest and sudden cardiac death.2 Therefore, patients with sinus bradycardia in the absence of heart disease are often advised to implant an electronic pacemaker for preventing unexpected sudden cardiac death because of a lack of pharmaceutical options.1
The sinus node generates a spontaneous pacing action potential. The diastolic potential in the sinus node is around −65 mV.3 HCN channels are highly expressed in the sinus node and are one of the main determinants in sinus node spontaneous pacemaker activity.3 HCN channel activation is voltage-dependent.4 HCN channel-generated inward current (also called cardiac pacemaker current, If) begins to activate around −45 to −50 mV in the sinus node5, 6 (defined as the threshold voltage of activation, Vth) and thus contributes to early diastolic depolarization in the sinus node.6
The HCN channel family has four members (HCN1-4).7 In mammalian expression cell lines, HCN1 channels exhibit the most positive threshold voltage activation (−45 to −50 mV) associated with the fastest activation kinetics (~100 milliseconds near the threshold voltage activation), whereas HCN4 has the most negative threshold voltage activation (around −65 mV) associated with the slowest activation kinetics (~2000 milliseconds near the threshold voltage activation).8, 9
The first HCN channel protein mutation linked to sinus bradycardia was HCN4-573x, reported in 2003.10 So far, 30 HCN4 mutations have been identified in sinus bradycardia patients11 and in patients with familial inappropriate sinus tachycardia,12 Brugada syndrome,13 ventricular fibrillation and left ventricular noncompaction,14 left bundle branch block,15 prolonged QT intervals, ST segment elevation, and torsade de pointes tachycardia,4, 16 as well as sudden infant death syndrome.17 All HCN4 mutations are heterozygous. All loss-of-function mutations caused a reduced HCN pacemaker current via a negative shift of voltage-dependent activation18 and/or decreased channel expression at the cell surface that reduces current density.10, 19, 20 Both mechanisms cause HCN channels to produce a decreased inward current at diastolic depolarization, which reduces the sinus node spontaneous action potential and thus, slows the heart rate.
HCN4 transcript is most abundant in the sinus node of animals (mouse, rat, rabbit, canine) and humans.21-25 However, recent studies of HCN channel protein expression levels have convincingly demonstrated that HCN1 protein levels are the highest followed by HCN4 in mouse26 and human sinus node.27 Mice lacking HCN1 developed severe sinus bradycardia and reduced cardiac output.26 The ionic current (If encoded by HCN genes) that contributes to sinus node pacemaker activity is reduced by ~40%.26
Using both candidate gene and next-generation exome sequencing approaches, we report, for the first time, a novel missense variant in the HCN1 gene in a patient with profound sinus bradycardia, with the absence of missense mutations in HCN4, HCN2, and other voltage-gated ion channels (Nav1.5, Cav1.2, Cav1.3, Cav3.1) known to cause sinus bradycardia.
2 METHODS (DETAILED METHODS ARE PROVIDED IN SUPPLEMENTAL MATERIALS)
2.1 Human studies
The study protocol and blood drawing procedures were reviewed and approved by West Virginia University Institutional Review Board (IRB1508782471). Written consent forms were sufficiently explained in detail to, agreed, and signed by study participants.
2.2 Electrocardiography of subjects
Heart rate was recorded by a standard 12-lead ECG from the patient (P), the patient's son (PS), and a family unrelated healthy volunteer (V).
2.3 Genomic DNA isolation, HCN4 amplification, and direct sequencing
Genomic DNA was isolated from P, PS, and V using Qiagen DNA isolation kit for blood sample (QIAamp DNA Blood Mini Kit). HCN4 exons were amplified using the primers specified in Table S1 and purified by gel electrophoresis. The WVU Genomic Core performed direct sequencing of HCN4 PCR products using sequencing primers described in Table S2.
2.4 Whole-exome sequencing and variation analysis
Whole-exome sequencing was performed on genomic DNA samples from P, PS, and V by Applied Biological Materials Inc (ABM). Variation-call-format (VCF) files were generated by ABM, then visualized and analyzed using Integrative Genomic Viewer28 and gnomAD29 for missense variations.
2.5 GFP-tagged HCN1 mutant constructs
Wild-type human HCN1 (WT or wtHCN1) in pcDNA3 was used for the construction of variants for 72GGG74 and P851A. wtHCN1 and mutants were then cloned into the N-terminus of pEGFP-C1 mammalian expression vector.
2.6 Cell culture and transfection
Human embryonic kidney (HEK293) cells were used for the expression of wild-type and mutant HCN1 channels. Transient plasmid transfection was performed using Lipofectamine_3000 (Invitrogen) following the vendor's instructions.
2.7 Western blotting
HEK293 cells transfected with wtHCN1 and mutant channel plasmids were harvested in RIPA buffer with fresh protease and phosphatase inhibitor cocktail (Sigma) on ice using a cell scraper. Detailed procedures are similar to our previous report9 and are provided in Data Supplements.
2.8 Flow cytometry
HEK293 cells expressing wtHCN1 and mutants were harvested 2 days after transfection using 0.25% trypsin with EDTA. Samples were analyzed in the WVU Flow Cytometry & Single Cell Core Facility on a four laser BD LSRFortessa. Details are provided in Data Supplements.
2.9 Confocal imaging of HEK293 cells expressing HCN1 mutant channels
HEK293 cells transfected with wild-type and mutant HCN1 channels were incubated on coverslips and fixed in 4% paraformaldehyde/PBS. The cells were imaged by the Nikon Sweptfield Confocal microscopy.
2.10 Whole-cell patch-clamp electrophysiology studies
Whole cell patch-clamp studies were carried out in HEK293 cells at 35-37℃. Details of patch-clamp studies are similar to our previous reports9, 30 and are provided in Data Supplements.
Threshold voltage (Vth) for HCN channel activation was defined as the least negative voltage at which HCN channels begin to open, generating a time-dependent inward current of at least 10pA was observed detected (a value that allowed clear discrimination from background noise, as seen in our previous publication31).
2.11 Sinoatrial pacemaker activity cell model
OpenCOR 0.1 software32 was used to simulate the effect of HCN1 variant changes in sinoatrial pacemaker activity using an updated sinoatrial pacemaker activity cell model.33
2.12 Statistical analysis
Statistical analyses were performed by one-way ANOVA followed by post hoc test Bonferroni (all means compared to wtHCN1) or Tukey (multiple comparisons with each other). The averaged data were presented as means ± SD. A P-value of less than.05 was considered as statistically significant.
3 RESULTS
3.1 Patient studies
The patient, a 42-year-old African American, was not on any medication nor had electrolyte abnormalities. Despite no evidence of structural heart disease by echocardiography, a 12-lead EKG showed profound symptomatic sinus bradycardia in his 30s. His mother had been previously diagnosed with sick sinus syndrome and underwent pacemaker implantation. The patient received a dual-chamber pacemaker and experienced complete relief of symptoms.
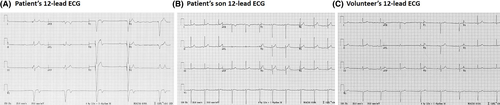
The patient's son was a healthy teenage boy (age 15 years), and a family unrelated healthy volunteer (male, 26 years) who is an amateur long-distance runner, served as a volunteer control. Resting heart rate was obtained from a 12-lead electrocardiograph (ECG) for P (Figure 1A, HR = 31 bpm, with complete heart block), PS (Figure 1B, HR = 67 bpm), and V (Figure 1C, HR = 50 bpm).
3.2 Direct sequencing of HCN4 gene
Previous studies have established HCN4 mutations as one of the genetic causes in patients with sinus bradycardia. HCN4 gene contains 8 exons.10, 34 We used amplification primers (Table S1) to obtain each exon from genomic PCR and subsequent gel electrophoresis, and sequencing primers (Table S2) to sequence the HCN4 gene. We found no missense mutations in HCN4 gene from the patient, the son, and the volunteer.
3.3 Exome-sequencing discovery of HCN1 variants in the patient
To explore a comprehensive genetic cause of the patient's bradycardia, we performed next-generation exome sequencing on genomic DNA isolated from P, PS, and V. In HCN4 gene, we found no missense variant, but did find one known heterozygous synonymous variant (rs529004) in the C-terminal region, shared by PS and V (Figure 2A). In HCN2 gene (another HCN isoform expressed at a very low level compared to HCN1 and HCN4 in the sinus node21, 24), we found no missense, but five known heterozygous synonymous variations (rs55659726, rs56170955, rs55780677, rs12981860, rs1054786).
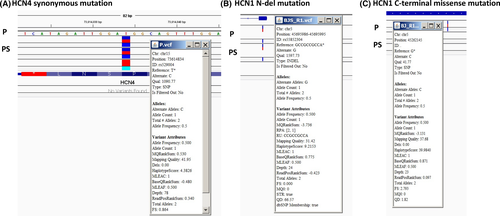
In HCN1, two variants were found in the patient. One is located in the N-terminus in HCN1 exon_1, rs55852304 (now merged into rs560604803), located in Human hg19: Chr5: 45695986-45695995. It is an in-frame deletion (indel), GCCGCCGCC, which encodes 72GGG74 in the protein sequence (HGVS nomenclature: NM_021072.3:c. 187_195delGGCGGTGGC). This 72GGG74 deletion is found in three gene variant databases mentioned above and is shared by the patient's son (Figure 2B). This allele frequency in global population according to 1000Genomes is 2.2% (https://www.ncbi.nlm.nih.gov/snp/rs56064803#frequency_tab).
A missense c.2551C>G transversion variant was found in Human hg19: Chr5: 45262145, in exon8 of HCN1, causing a p. Pro851Ala [CCC (Pro) → GCC(Ala)] missense mutation (Figure 2C). The protein and nucleotide sequences alignments are shown in Figure S1 and S2). This variant is not found in (1) National Heart, Lung, Blood Institute (NHLBI) GO Exome Sequencing Project (ESP) [Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (http://evs.gs.washington.edu/EVS/) [accessed: Jan 11, 2019], containing 6403 samples; (2) Exome Aggregation Consortium (ExAC) (which has about 60 000 individuals) (http://exac.broadinstitute.org/) [accessed: Jan 11, 2019]; and (3) the genome Aggregation Database (gnomAD, an upgraded version of ExAC, covering two datasets: exome sequence data from 123 136 individuals and whole genome sequencing from 15 496 individuals, https://gnomad.broadinstitute.org/gene/ENSG00000164588) [accessed: Jan 12, 2019] (Figure S3). To our best knowledge, P851A is a novel variant that has not been reported before in bradycardia patient.
Sequencing alignment across species on MARRVEL35 indicated that P851 is conserved in the polypeptide sequence of HCN1 of human (hs, Homo sapiens), mouse (mm1, Mus musculus), rat (rn1, Rattus norvegicus), Zebrafish (dr1, Danio rerio), and Fly (dm1, Drosophila melanogaster) (arrows in Figure S4). PolyPhen-236 predicted that P851A variant is “probably damaging” with a score of 0.999. The Sorting Intolerant From Tolerant (SIFT) algorithm predicts the effects of a missense variant on protein function.37, 38 SIFT predicted that P851A is not tolerated (Supplemental excel file HCN1 SIFT, row 62, highlighted).
Missense mutations in the voltage-dependent calcium (Cav1.2, Cav1.3, Cav3.1) and sodium (Nav1.5) channels have been reported to be linked to bradycardia.39-42 We found no missense mutations in these channel genes SCN5A (Nav1.5), CACNA1C (Cav1.2), CACNA1D (Cav1.3), CACNA1G (Cav3.1, T-type) from the patient. We have also studied sequences of other genes associated with bradycardia (KCNK17, RYR2, CASQ2, AMK2, MYH6),43 and found no missense mutations.
From VCF (variant call format) files, the patient was heterozygous for N-del and P851A. The patient's son has the N-del but not the P851A. Although additional blood samples from the patient's parents and siblings were unavailable, it is almost certain that the two mutations are on different alleles, otherwise, the son would inherit both or neither. Thus, we studied co-expression of N-del + P851A (mimic the patient's variant) and N-del + wtHCN1 (mimic the son's variant).
3.4 Protein expression of HCN1 mutants
We first examined the surface expression of HCN1 mutants transfected into HEK293 cells. In membrane preparations, a human HCN1 antibody detected protein expression of WT, N-del, and P851A HCN1 channels at the expected molecular weight (~120 kDa) (Figure 3A). Co-expression of wide-type HCN1 with the mutant channels (N-del + wtHCN1 for PS, N-del + P851A for P) are shown in Figure 3B.
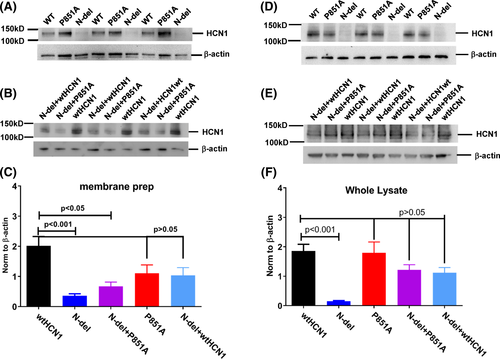
On average, there was over five-fold reduction in surface expression of N-del, compared to the wild-type HCN1 channel protein, after being normalized to the loading control (Figure 3C) (WT: 2.01 ± 0.830, n = 7; N-del: 0.36 ± 0.17; n = 6, P < .001). There was a three-fold reduction of surface expression in N-del + P851A compared to the wildtype channel (WT: 2.01 ± 0.830, n = 7; N-del + P851A: 0.67 ± 0.28; n = 4, P < .01). There was no statistical difference in surface expression levels of P851A comparing to the wildtype channels (WT: 2.01 ± 0.830, n = 7; P851A: 1.11 ± 0.55; n = 4, P > .05) and N-del + wtHCN1 (WT: 2.01 ± 0.830, n = 7; N-del + wtHCN1: 1.04 ± 0.44; n = 4, P > .05) channel proteins (Figure 3C).
Total channel protein levels were examined in the whole lysate preparations. N-del (Figure 3D) and N-del + P851A (Figure 3E) showed a reduction in channel protein levels compared to wildtype HCN1. The channel protein levels are decreased by ~12-fold for N-del in comparison to the WT, while changes in P851A, N-del + P851A, and N-del + wtHCN1 were statistically not significant (Figure 3F) (WT: 1.86 ± 0.56, n = 6; N-del: 0.15 ± 0.04, n = 6; P851A: 1.80 ± 0.81, n = 5; N-del + P851A: 1.21 ± 0.44, n = 6; N-del + HCN1wt: 1.12 ± 0.40, n = 5).
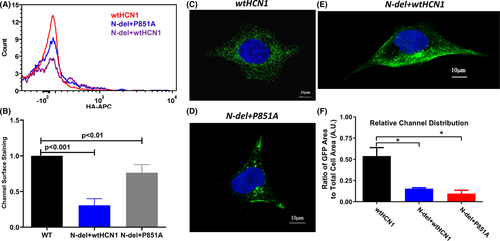
Additionally, we used flow cytometry to detect the surface expression of channel proteins at the single cell level. A hemagglutinin (HA) tag in the extracellular loop between S3 and S4 transmembrane domains was introduced into the HCN1 wild-type and mutant constructs (WT, P851A, N-del). We used an HA-antibody conjugated with Alexa647 to stain surface expression of HCN1 mutants in cells without permeabilization. For intracellular staining, we used an HCN1-antibody conjugated with phycoerythrin (PE) fluorophore after cells were permeabilized. The HCN1 antibody targets an intracellular epitope (amino acid residues 790-883) in the C terminus of the protein (Thermo Scientific, PA553950). GFP-positive cells that bind to HA-APC antibody prior to permeabilization were counted (Figure 4A). The fluorescent signals are contributed by HA-tagged HCN1 channels expressing at the cell surface. The total number of channels at cell surface is the product of the count and the median or geometric mean of HA-APC fluorescent signals (Figure 4B). Surface expression of N-del + wtHCN1 channels are decreased by 69% when normalized to WT (WT: 1, N-del + wtHCN1: 0.31 ± 0.09; n = 5, P < 001). Expression levels of N-del + P851A channels at cell surface are also decreased, but to lesser extent, 24% (WT: 1; N-del + P851A: 0.76 ± 0.11; P < .01, n = 5). There is no statistical difference between the surface expression levels of P851A and WT HCN1 channels (WT: 1; P851A: 0.92 ± 0.17; P > .05, n = 5). Figure S5 shows that surface expression of N-del channels is decreased by 71% (WT: 1; N-del: 0.29 ± 0.11; P < .001, n = 5), but the surface expression of P851 channels is not statistically different from wild-type channel (WT: 1; P851A: 0.87 ± 0.31; P > .05, n = 5).
3.5 Confocal imaging studies of HCN1 mutants
We also performed confocal imaging studies of HCN1 mutant channels co-expressed with the wild-type HCN1 in HEK293 cells. Representative channel distributions (green) are shown for the volunteer (wtHCN1) (Figure 4C), the patient (N-del + P851A) (Figure 4D), and the patient's son (wtHCN1 + N-del) (Figure 4E). The relative channel distribution was estimated by calculating the total number of the expressed channels divided by the total cell area from the brightfield image, the averaged results are shown in Figure 4F. Compared to the wild-type HCN1, the total number of channels was >fivefold decreased for wtHCN1 + P851A and >3-fold decreased for wtHCN1 + N-del (wtHCN1: 0.54 ± 0.20, N-del + wtHCN1: 0.16 ± 0.02, N-del + P851A: 0.095 ± 0.079, n = 6, P < .05). There was no significant difference in the average ratio of GFP area to total cell area between N-del + P851A and N-del + wtHCN1 using one-way ANOVA followed by post hoc test Tukey (N-del + wtHCN1: 0.155 ± 0.0593, N-del + P851A: 0.0958 ± 0.0884, P > .05) (Figure 4F).
3.6 Biophysical properties of HCN1 mutants
Studies of HCN4 mutations have revealed two mechanisms by which missense mutations cause reduced channel activity: reduced channel expression and/or a negative shift of voltage-dependent activation.18 Therefore, we investigated the threshold activation (the least negative potential at which the HCN channel inward current of 10pA can be detected31) and the current density (the whole-cell current divided by cell capacitance), which reflects the functional channels of HCN1 mutants available at the cell surface.
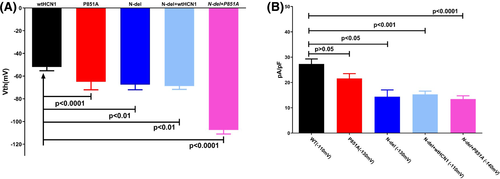
Figure 5 shows the representative current activation of wild-type human version HCN1 (wt-hHCN1) and the mutant channels expressed in HEK293 cells. Channel-expressing cells were voltage-clamped at −20 mV and hyperpolarized to a range of potentials marked in the figure. Compared to wild-type HCN1 (Figure 5A), N-del (Figure 5B), P851A (Figure 5C), N-del + P851A (Figure 5D), and N-del + wtHCN1 (Figure 5F) mutant channels were activated at more negative potentials. The average threshold activation is −52.0 ± 3.3 mV (n = 7) for WT, −65.0 ± 7.1 mV (n = 10) for P851A, −67.5 ± 4.5 mV (n = 10) for N-del, −68.8 ± 8.3 mV (n = 7) for N-del + wtHCN1, and −107.5 ± 12.2 mV (n = 12) for N-del + P851A, respectively (Figure 6A). There exists a statistically significant negative shift in the threshold activation for P851A (−13 mV), for N-del (−15 mV), for N-del + wtHCN1 (−16 mV), and for N-del + P851A (−55 mV), respectively, when compared to the wild-type HCN1 (Table 1). There was a significance difference of HCN1 mutant channel threshold activation between N-del + P851A and N-del + wtHCN1 using one-way ANOVA followed by post hoc test Tukey (N-del + wtHCN1: −68.75 ± 8.35 mV, N-del + P851A: −107.5 ± 12.15 mV, P < .05) (Figure 6A).
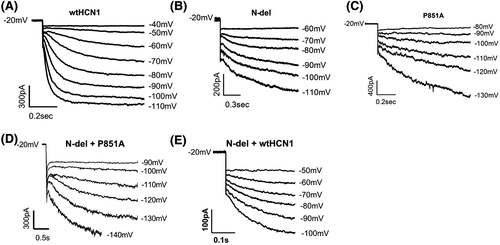
| WT | N-del | P851A | N-del + P851A | N-del + HCN1wt | |
|---|---|---|---|---|---|
| Vth (mV) | −52.0 ± 3.3 (n = 7) | −67.5 ± 4.5* (n = 10) | −65.0 ± 7.1*** (n = 10) | −107.5 ± 12.2*** (n = 12) | −68.8 ± 8.3** (n = 7) |
| V50 (mV) | −68.8 ± 2.3 (n = 6) | −87.70 ± 4.6** (n = 7) | −91.56 ± 3.7** (n = 7) | −118.30 ± 15.1*** (n = 8) | −81.1 ± 7.6* (n = 5) |
| τVth (ms) | 98.3 ± 38.2 (n = 7) | 468.1 ± 79.5** (n = 10) | 353.4 ± 50.7** (n = 10) | 925.2 ± 311.8** (n = 8) | 124.8 ± 30.8 (n = 6) |
| pA/pF (max. mV) | 27.3 ± 2.0 (−110 mV) (n = 5) | 14.4 ± 2.7* (−130 mV) (n = 7) | 21.6 ± 1.9 (−130 mV) (n = 6) | 13.5 ± 3.4*** (−140 mV) (n = 7) | 15.3 ± 3.3*** (−110 mV) (n = 5) |
- * P < .05 compared to WT
- ** P < .01 compared to WT
- *** P < .001 compared to WT.
The half-voltage activation of mutant channels is also significantly shifted to hyperpolarizing potentials compared to wild-type (Table 1): N-del by −19 mV, P851A by −23 mV, N-del + P851A by −49 mV, and N-del + HCN1wt by −12 mV, respectively. Wild-type HCN1 channel has fast activation kinetics (100-300 milliseconds near threshold voltage of activation).8, 44 All mutant channels have slower activation kinetics near their respective threshold voltage of activation (τVth) (Table 1), notably, N-del + P851A has the slowest activation kinetics near the voltage threshold of activation.
We next measured the current density (current amplitude normalized to the cell capacitance, pA/pF) of HCN1 mutant channels at the voltage where the channels are fully activated to calculate the relative functional channels available at the cell surface. Compared to WT, there is a 47% reduction of current density for N-del (WT (−110 mV): 27.3 ± 2.0 pA/pF, n = 5; N-del (−130 mV): 14.4 ± 2.7 pA/pF, n = 7), a 51% reduction of current density for N-del + P851A (WT (−110 mV): 27.3 ± 2.0 pA/pF, n = 5; N-del + P851A (−140 mV): 13.5 ± 3.4 pA/pF, n = 7), and a 44% decrease in current density for N-del + HCN1wt (WT (−110 mV): 27.3 ± 2.0 pA/pF, n = 5; N-del + HCN1wt (−110 mV): 15.3 ± 3.3 pA/pF, n = 5). On the other hand, P851A showed no statistical decrease in current density compared to WT HCN1 channels (WT (−110 mV): 27.3 ± 2.0 pA/pF, n = 7; P851A (−130 mV): 21.6 ± 1.9 pA/pF, P > .05, n = 8) (6B, Table 1). There was no significance difference of HCN1 mutant channel current density between N-del + P851A and N-del + wtHCN1 using one-way ANOVA followed by post hoc test Tukey (N-del + wtHCN1: 15.33 ± 3.33 pA/pF, N-del + P851A: 13.47 ± 3.39 pA/pF, P > .05) (Figure 6B).
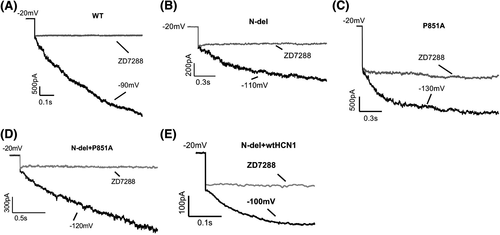
One of the hallmarks of HCN channels is the functional channel blockade by ZD7288, a selective blocker for HCN pacemaker channels.45 Figure 7 shows the ionic current inhibition of HCN1 wild-type (A), mutant channels (B, N-del; C, P851A), co-expression of N-del and P851A (D), and co-expression of N-del with wild-type channel (E) by 10 μM ZD7288 at the respective membrane potential.
3.7 Effect of HCN1 variant in the sinoatrial pacemaker activity
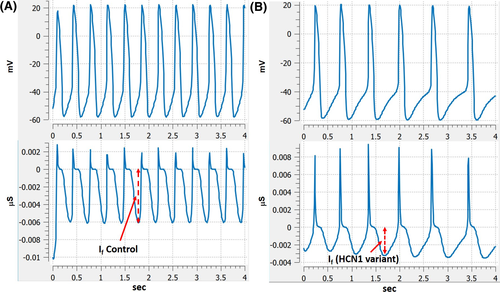
To evaluate the effect of HCN1 variant in the sinoatrial pacemaker activity, we employed an updated sinoatrial pacemaker activity cell model developed by DiFrancesco's group.33 HCN1 protein levels are >15-fold higher than HCN4 and HCN2 in the sinus node.27 We assumed that removing the main contributor HCN1 result in one-third decrease in I(f) channel conductance; the simulation result showed that the rate of spontaneous action potential is decreased by about 50% because of reduction in the initial phase 4 depolarization speed (control: ~110 mV/s, HCN1 variant: ~60 mV/s) (Figure 8, A-control action potential (upper) and I(f) (lower), B-reduced rate of action potential (upper) and I(f) (lower)).
4 DISCUSSION
In the index patient, both direct- and exome-sequencing revealed no missense mutations in HCN4; instead, we found two variants in the HCN1 gene. One variant is an indel located in the N-terminus (72GGG74) and the other is P851A located in the C-terminus. The 72GGG74 N-del is found in the National Heart, Lung, Blood Institute GO Exome Sequencing Project database [Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/)].
Currently, 29 variations in HCN1 gene have been found in patients with epilepsy.46 Four (T171R in the S1-S2 loop, C329S in the S5-S6 loop, V414M and S680Y in the C-terminus) are characterized as inherited, likely pathogenic variations, nineteen are novel pathogenic variants, and the rest are of unknown clinical significance. N-del (72GGG74 indel, p. Gly72_Gly74del, or c.187_195del) was previously found in idiopathic generalized epilepsy (rs56064803)47 and in an early infantile epileptic encephalopathy patient, but its clinical significance has been characterized as benign using a Sherloc criteria48 (National Center for Biotechnology Information. ClinVar; Variation ID 95999, https://preview.ncbi.nlm.nih.gov/clinvar/variation/95999/ (accessed Feb. 24, 2019)). To our best knowledge, this is the first time that a patient exhibiting the rs56064803 indel with profound sinus bradycardia has presented with absence of epilepsy.
P851A (c.2551C>G) is not found in NHLBI GO exome and gnomAD databases and has not been previously reported in the literature. Therefore, we consider it a de novo missense variant in HCN1 gene. Protein expression studies using immunoblots, confocal imaging, and flow cytometry indicated that N-del is defective in biosynthesis, resulting in reduced channel expression at the cell surface. On the other hand, P851A does not exhibit a significant change in channel surface expression compared to the wild-type channels.
Co-expression of N-del with P851A also showed decreased cell surface expression of the channels, although the total protein synthesis was not statistically reduced compared to the wild-type channel. Decreased surface expression of co-expressed N-del with wild-type HCN1 (patient's son situation) was not detected by immunoblots, but by confocal imaging and flow cytometry. Importantly, the decreased number of functional channels of N-del + wtHCN1 were reflected in patch-clamp studies as significantly reduced current density.
Alterations in voltage-dependent activation of the mutant channels are of direct relevance to the inward current contributed to the early diastolic depolarization phase in sinus node action potential. Both N-del and P851A have a negative shift in the threshold voltage activation associated with a simultaneous, significant slowing of activation kinetics, compared to the wild-type HCN1. The compound variant, N-del + P851A, has the largest hyperpolarizing shift in the threshold voltage activation versus N-del and P851A. Additionally, among the three HCN channel isoforms present in the sinus node, HCN1 has the fastest activation kinetics during the diastolic phase, comparable to pacemaker current activation kinetics in the sinus node (~1s).49 A combination of negative shift of voltage-dependent activation and a significantly slowed activation kinetics make the compound mutant (N-del + P851A) channel unable to contribute to the diastolic depolarization of the sinus node. This channel gating mechanism also explains why the patient's son is asymptomatic: although surface expression of N-del + wtHCN1 is reduced, the channel activities (Vth, V1/2, and τth) are not suppressed to the extent seen in the compound variant (N-del + P851A).
To further understand how the compound mutant (N-del + P851A) affects the heart rate in the patient, we performed a simulation of sinoatrial pacemaker activity affected by HCN1 variant. The simulation result demonstrated that removal of HCN1 contribution to overall pacemaker current significantly decreased pacemaker channel conductance, resulting in a decreased phase-4 depolarization and a nearly 50% reduction in the pacemaker activity, which mimics the profound bradycardia in the indexed patient.
5 CONCLUSIONS
In the present work, we report two HCN1 variants found in the indexed bradycardia patient, in the absence of missense mutations of HCN4 and other channels known to cause cardiac bradycardia. Our data indicated that N-del HCN1 has defective protein biosynthesis, but unlikely contributes to the patient's bradycardia since it is shared by his son who does not have bradycardia. The novel P851A variant does not have reduced channel expression at cell surface but exhibits a negative shift of voltage-dependent activation. The combination of two variants, N-del + P851A (present only in the patient), has the largest negative shift of voltage-dependent activation of HCN1 channels beyond the early diastolic depolarization of sinus node, which represents the likely cause of profound sinus bradycardia in the patient in the absence of missense variants in other ion channels previously linked to sinus bradycardia.
ACKNOWLEDGMENTS
The authors would like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010). The human version of wild-type HCN1 cDNA plasmid was kindly provided by Dr Juliane Stieber (Institut Für Experimentelle Und Klinische Pharmakologie Und Toxikologie Friedrich - Alexander Universität Erlangen, Germany). We thank WVU Genomic Core for PCR sequencing of HCN4 gene. We are also grateful for assistance from research nurse Annina Guzek for IRB process and blood drawing.
CONFLICT OF INTEREST
Authors declare no conflict of interest for this article.




