Expression of individual mutations and haplotypes in the galactocerebrosidase gene identified by the newborn screening program in New York State and in confirmed cases of Krabbe's disease
SIGNIFICANCE Newborn screening for Krabbe's disease has been instituted in several states. Individuals with galactocerebrosidase (GALC) activity below a cutoff level have received additional testing, including performance of molecular analysis. Several variants that had not previously been seen in confirmed patients have been identified. In some cases, polymorphisms have been seen in cis with these variants. These variants were expressed in COS1 cells, and the GALC activity measured was compared with known mutations and normal sequence. In some cases, the presence of polymorphisms on the same copy of the gene as a mutation resulted in very low GALC activity. The information obtained will be useful for counseling families of screen-positive newborns found to have low GALC activity.
Abstract
Newborn screening (NBS) for Krabbe's disease (KD) has been instituted in several states, and New York State has had the longest experience. After an initial screening of dried blood spots, samples from individuals with galactocerebrosidase (GALC) values below a given cutoff level were subjected to additional testing, including sequencing of the GALC gene. This resulted in the identification of mutations that had previously been found in confirmed KD patients and of variants that had never previously been reported. Some individuals had variants considered to be polymorphisms, alone or on the same allele as another mutation. To help with counseling of families on the risk for a newborn to develop KD, expression studies were conducted with these variants identified by NBS. GALC activity was measured in COS1 cells for 140 constructs and compared with mutations that had previously been seen in confirmed cases of KD. When a polymorphism was present on the same allele as the variant, expressed activity was measured with and without the polymorphism. In some cases the presence of the polymorphism greatly lowered the measured GALC activity, possibly making it disease causing. Although it is not possible to predict conclusively whether a variant is severe and will result in infantile KD if two such variants are present or whether a variant is mild and will result in late-onset disease, some variants clearly are not disease causing. This is the largest expression study of GALC variants/mutations found in NBS and confirmed KD cases. This work will be helpful for counseling families of screen-positive newborns found to have low GALC activity. © 2016 Wiley Periodicals, Inc.
Krabbe's disease (KD), or globoid leukodystrophy, is an autosomal recessive disorder caused by a deficiency of galactocerebrosidase (GALC) activity (Suzuki and Suzuki, 1970). GALC activity is required for the lysosomal hydrolysis of certain galactolipids, including galactosylceramide (galactocerebroside), psychosine, monogalactosyl diglyceride, and under certain conditions lactosylceramide (for review see Wenger et al., 2013). The deficiency of this enzyme results in defects in the production of healthy, stable myelin, leading to a variety of clinical and pathological features, depending on the onset of disease and time course. Although many patients have disease onset before 6 months of age (infantile form), other patients present later, from the late-infantile period to adulthood (Wenger et al., 2013). Although it can be surmised that patients who have a later onset have a small amount of residual GALC activity, it is difficult to measure this accurately in the tissue samples available. It has been proposed that onset of clinical features in later-onset patients is precipitated by some environmental event, such as infection or trauma to the head (Wenger, 2011; Debs et al., 2013). Most individuals who present with some clinical features undergo laboratory testing to arrive at a diagnosis, but by this time the disease would have started to progress, so treatment options would be limited. To obtain a diagnosis before any clinical features appear, newborn screening (NBS) for KD has been instituted in several states (Duffner et al., 2009; Orsini et al., 2016). New York State has screened over 2 million newborns since 2006; other states have only recently started screening. Testing involves the use of dried blood spots for initial screening, followed by molecular analysis and conventional testing for GALC activity for those who have GALC activity below a given cutoff level. NBS has identified some individuals who clearly would have an infantile-onset disease as well as some who may present when older, as determined by the presence of mutations previously found in patients with later onset. Some individuals who show low GALC activity in dried blood spot testing may have activity-lowering polymorphisms alone or together with one copy of a disease-causing mutation; these individuals will not have KD at any age.
After the cloning of the GALC cDNA and gene, molecular testing of confirmed patients with KD could be instituted (Chen et al., 1993, Luzi et al., 1995). In addition to the more than 147 mutations that have been identified in patients of all ages, other nucleotide changes considered polymorphisms have also been identified (Wenger et al., 2013). Some mutations found in patients can be considered mild, and others can be considered severe because of the clinical presentation in multiple patients. Sometimes what are considered disease-causing mutations occur in cis with mutations that are considered polymorphisms. In fact, the presence of a polymorphism in cis with another mutation may render the allele pathogenic (Wenger et al., 2014). Identification of mutations in patients began soon after the cloning of the GALC gene. These mutations were introduced into the normal GALC cDNA, and the resulting plasmids were usually placed in COS cells for expression studies. This was done to confirm the deleterious nature of a mutation. At times, a mutation in question was expressed with and without a known polymorphism that was present in that allele of the patient. This resulted in interesting and important findings. After the institution of NBS in New York, which includes molecular analysis of specimens with low GALC activity, additional mutations that had not previously been found in confirmed cases of KD have been identified (Orsini et al., 2016). It is critical to the future management of these individuals that attempts be made to determine whether such variants, when expressed, give very low or significant GALC activity. In an attempt to answer this question, many mutations found in known patients as well as novel variants identified in NBS were expressed in COS1 cells. When possible, expression of mutations was measured with and without the polymorphism(s) present on the allele, which was determined by parental testing for phasing. Variants that were expected to be disease causing because they yielded very low GALC activity after laboratory testing, such as small deletions and insertions resulting in a frame shift, large deletions, and premature stop codons, were tested as well.
MATERIALS AND METHODS
Normal and Mutant GALC cDNA Cloning
The New York State Department of Health NBS laboratory received the normal GALC cDNA clone from the lysosomal diseases testing laboratory at the Thomas Jefferson University, Philadelphia. The GALC clone is on the eukaryotic expression vector pcDNA3 (Invitrogen, Carlsbad, CA), prepared as described elsewhere (Luzi et al., 1996), and was sequence verified to be normal with only the sequence after the initiation codon changed to provide the Kozak sequence (RCCATGG; Chen et al., 1993). It should also be noted that the numbering system of the GALC cDNA and protein used in this article is the original one reported (Chen et al., 1993; Wenger et al., 2013).
Site-Directed Mutagenesis
The Agilent Technologies (Santa Clara, CA; formally Stratagene) QuikChange II site-directed mutagenesis kit was used to insert mutational and polymorphic changes to the GALC gene clone, according to the manufacturer's instructions. Briefly, the reaction consisted of 1.25 μl 10 × buffer, 0.25 μl PFU-Taq polymerase, 0.25 μl dNTP, 0.325 μl F-primer, 0.325 μl R-primer, and 9.1 μl molecular-grade H2O for a total volume of 11.5 μl. One microliter of GALC gene clone plasmid (100 ng) was added to the reaction as the template. Mutagenesis program parameters were, step 1, 95 °C for 30 sec; step 2, 95 °C for 30 sec; step 3, 54 °C→64 °C gradient for 1 min; step 4, 68 °C for 10 min, cycling back to step 2 for 14 more cycles; step 5, 25 °C for 10 min. The mutagenesis reactions were performed on a Veriti 96-well Thermal Cycler (Life Technologies, Grand Island, NY). Afterward, 6 μl of the mutagenesis reactions was electrophoresed on a 1% agarose gel to check for adequate amplification prior to transformation. The best reactions were then digested to remove any normal sequence template with 1 μl Dpn1 and incubated at 37 °C for 60 min. One microliter was then used for transformation into competent Escherichia coli cells.
Transformation
XL1-Blue supercompetent cells from the QuikChange II kit were transformed with the mutant clones. The transformations were performed according to the protocol provided with the QuikChange II kit. Individual colonies were cultured in lysogeny broth media plus 100 mg ampicillin for selection and allowed to grow for 24 hr. Cultures were then centrifuged at 1,500 rpm for 15 min. The Omega Bio-Tek (Norcross, GA) Plasmid Mini Kit I was used to isolate the mutated GALC clone plasmids.
Sequencing
The concentration of each clone was determined with a Thermo Scientific (Wilmington, DE) NanoDrop 1000 spectrometer. Each clone was diluted to a concentration of 100 ng/μl. All clones were Sanger sequenced (primers available on request) on a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA) to ensure that only the correct, expected sequence was present. Clones were numbered and stored at –20 °C until used for expression studies. Expression studies were conducted by the lysosomal diseases testing laboratory at Thomas Jefferson University.
Transfection and GALC Activity Determination
COS1 cells were seeded in six-well plates on the day before the transfection. Cells at approximately 50% confluence were transfected with Attractene transfection reagent (Qiagen, Germantown, MD). Briefly, the transfection complex mix was prepared for each well by mixing 12 µg plasmid DNA in a final volume of 100 µl in DMEM with 4.5 µl Attractene reagent. The complex was mixed by vortexing, centrifuged in a microfuge for a few seconds, and incubated at room temperature for 15 min to allow for complex formation. In the meantime, 2 ml fresh complete medium (DMEM + 10% fetal bovine serum and antibiotics) was added to each well. The transfection complexes were then added dropwise onto the cells while swirling the plate to ensure uniform distribution of the complexes. The cells were then incubated for 72 hr under normal growing conditions (37 °C and 5% CO2).
After 72 hr, the cells were harvested, washed with phosphate-buffered saline, resuspended in 200 µl water, and sonicated with a probe-type sonicator tip, and the protein concentration was determined (Lowry et al., 1951). GALC activity was measured with [3H]galactosylceramide substrate, as previously described by Wenger and Williams (1991).
All clones containing mutations in the GALC gene were transfected at least in duplicate with the exception of the clones containing deletions and insertions, which were transfected only once. Untransfected COS1 cells (mock) had an average GALC activity of 1.0 nmol/hr/mg protein (range 0.8–1.3 nmol/hr/mg protein in 14 transfection studies). The average activity in the cells transfected with the GALC clone containing no mutations (WT) was 24.5 nmol/hr/mg protein, with a range of 18.0–29.9 nmol/hr/mg protein from 19 separate transfections.
RESULTS
All mutations were introduced independently into pcDNA3 containing WT GALC. Haplotypes were built by consecutive introduction of known and/or NBS-identified variants in cis. GALC mutations that are expected to have little to no GALC activity (deletions, insertions, nonsense mutations) are shown in Figure 1. GALC activities in the cells transfected with the mutations of interest were compared with COS1 cells transfected with a plasmid containing normal sequence (WT) and with untransfected COS1 cells (mock). Just as predicted, none of these mutations resulted in GALC activities above the untransfected COS1 cells. Asterisks in Figure 1 indicate mutations that have also been expressed in cis with p.I546T, a polymorphism known to reduce the measured GALC activity partially. In these constructs, the presence of the polymorphism did not further reduce the low activity measured (not shown).
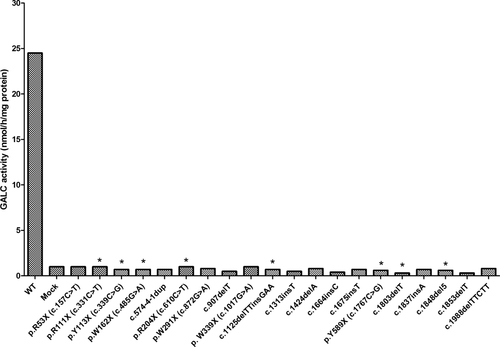
Expression of small deletions, insertions, and nonsense mutations in COS1 cells. Transfections and assays were performed as described in Materials and Methods. Mutations tested are indicated below each bar and include WT and untransfected cells (mock). Asterisks indicate mutations found in cis with p.I546T. Expression levels with or without this polymorphism were the same (not shown).
Figure 2A,B shows expression of mutations and haplotypes identified in patients with a diagnosis of KD. The red bars indicate the activities of the haplotypes found in the patients. The letter M or S above a bar indicates mild or severe, respectively. This distinction is noted in cases in which the genotype has been identified in patients with a phenotype of infantile (severe) or later-onset (mild) forms of KD (Wenger et al., 2013). No letter above a column indicates the severity of the mutation cannot be predicted. It is of note that both Figure 2A and Figure 2B show that enzyme activity is reduced in constructs in which one or more polymorphisms are in cis with a mutation. This “haplotype effect” is well demonstrated on the p.G41S + p.I546T, p.A44T + p.R168C + p.I546T, p.R63H + p.I546T, and p.D171V + p.R168C + p.I546T alleles (Fig. 2A) and on the p.M309V + p.I546T, p.Y319C + p.I546T, p.Y474N + p.I546T, p.R515C + p.I546T, p.L618S + p.I546T, and p.V665M + p.I546T alleles (Fig. 2B).
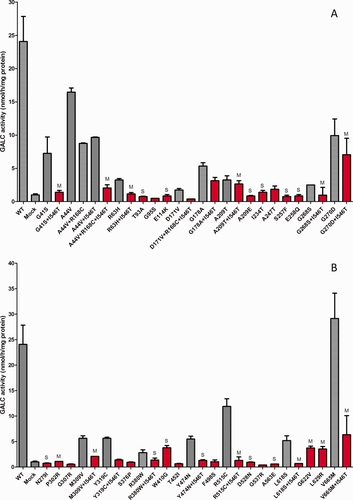
A,B: Expression of mutations and haplotypes found in patients confirmed to have KD. Transfections and assays were performed as described in Materials and Methods. Mutations tested are indicated below each bar and include WT and untransfected cells (mock). Red bars indicate the haplotype as found in patients, and gray bars indicate the mutation either alone or with other polymorphisms. The letter M or S above a bar indicates a mutation that is predicted to be mild or severe, respectively. Neither M nor S above a bar indicates that the mutation severity could not be predicted. All clones were transfected at least in duplicate. Mean and SD.
Figure 3A,B shows GALC activities of variants and haplotypes identified by NBS for KD in New York State. Red columns indicate actual haplotypes. Some were found in only one individual, and others were found in more than one. The haplotype effect is also remarkable for these alleles, e.g., p.R53Q + p.I546T, p.E60K + p.I546T, p.R63C + p.I546T, and p.P73L + p.I546T (Fig. 3A) and p.L634P + p.I546T (Fig. 3B). Although the presence of polymorphisms in cis often further decreases the measured GALC activity when present with most variants, the presence of some polymorphisms may have no effect on the measured activity (e.g., p.G399R; Fig. 3B) or in fact may raise the measured activity (e.g., p.M101V; Fig. 3A). Finally, it is obvious from the measured activities that some variants identified by NBS would not be disease causing whether a polymorphism was present or not (e.g., p.N151S + p.I546T, p.V320M + p.I546T, and p.T452S + p.A5P + p.G9G + p.D232N; Fig. 3B).
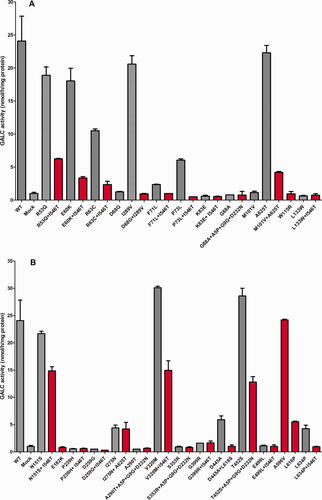
A,B: Expression of mutations and haplotypes found in newborn screening. Transfections and assays were performed as described in Materials and Methods. Mutations tested are indicated below each bar and include WT and untransfected cells (mock). Red bars indicate the haplotype as found in referred individuals, and gray bars indicate the mutation either alone or with other polymorphisms. All clones were transfected at least in duplicate. Mean and SD.
Two variants, p.Y303C and p.T96A, have been found in relatively high frequency in New York State NBS. They have also been found in patients with late-onset KD only when they occur in trans with certain second severe mutations (Wenger et al., 2013, 2014). Also, p.T96A has been identified in the heterozygous state in only two adult-onset KD patients. One patient has four additional polymorphisms in cis with p.T96A (Luzi et al., 1996), and the other is in a 1637T>C (p.I546T) polymorphic background (Debs et al., 2013). The effects of polymorphisms on the measured GALC activity on p.T96A and p.Y303C are shown in Figure 4. It is interesting to note that both p.T96A and p.Y303C are always found with three other polymorphisms, p.A5P, p.G9G, and p.D232N. However, the one confirmed late-onset patient with p.T96A also has p.I546T on the same allele. In over 2 million NBS samples, p.T96A has never been found in cis with p.I546T. It is interesting to note that the severe mutations p.R53X and p.R204X (Fig. 1) were identified by NBS as compound heterozygotes with p.T96A, but p.I546T was not present. In confirmatory testing of these individuals, the GALC activity was in the no- to moderate-risk range. In NBS, the c.1664insC (p.D556X) mutation (Fig. 1) was in trans with the p.Y303C haplotype (Fig. 4), and confirmatory testing showed GALC activity in the high-risk category. We predict that this individual will not have infantile KD but may develop a later-onset form.
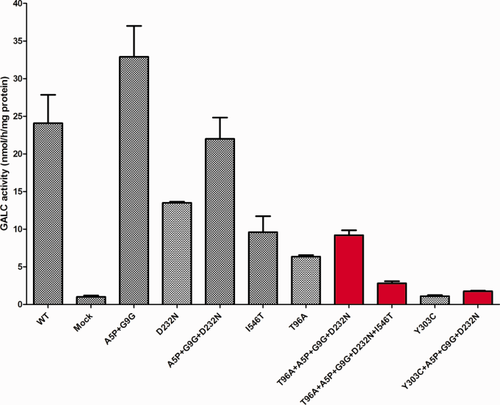
Expression of p.T96A and p.Y303C mutations with and without certain polymorphisms. Transfections and assays were performed as described in Materials and Methods. Red bars indicate the haplotype as found in confirmed patients and referred individuals, and gray bars indicate the mutations either alone or together in various combinations. All clones were transfected at least in duplicate. Mean and SD.
DISCUSSION
New York State instituted NBS for KD in 2006 (Duffner et al., 2009; Orsini et al., 2016). As of August, 2015, more than 2.2 million infants had been tested, 712 had been subjected to molecular analysis, and 393 had been referred for confirmatory analysis. Since the cloning of the GALC cDNA (Chen et al., 1993), about 150 mutations and several polymorphisms (normal allelic variants that do not result in disease when homozygous or in trans with a disease-causing mutation) have been identified. Newborns with a GALC value identified by NBS below an established cutoff level were subjected to additional testing, including sequencing of the GALC gene (Duffner et al., 2009). Sequence analysis identified mutations that had previously been reported in KD patients as well as novel variants that had not previously been detected in KD patients. Because of the multiple variants identified in some infants, it became evident that GALC molecular phasing was required to identify which variants (known, novel, or polymorphic) were on the same, in cis, or on the opposite, in trans, chromosome to facilitate clinical evaluation, correlation with confirmatory enzyme activity, followup, and genetic counseling. This is essential when novel variants or variants of unknown significance (VOUS) on particular genetic backgrounds are being interpreted. Another level of complexity was added by the high frequency of three well-known GALC polymorphisms (p.R168C, p.D232N, and p.I546T) found in patient and newborn populations. Among the referred population (detected by NBS), the carrier/allele frequencies were 0.589/0.359 (p.R168C), 0.420/0.246 (p.D232N), and 0.874/0.579 (p.I546T; Orsini et al., 2016). To the best of our knowledge, New York State has the largest available NBS database on known and novel (VOUS) GALC gene variants and their chromosomal haplotypes.
GALC expression studies have previously been performed (Luzi et al., 1996; De Gaspieri et al., 1996), in some cases with limited haplotype information. Mutations ideally should be studied in the same allelic background present in each individual. In an attempt to predict which of the variants identified by NBS might be disease causing, GALC expression studies have been conducted for individual mutations and their haplotypes.
To evaluate the capability of the method for identifying disease-causing mutations, expression studies of known and recently identified obviously deleterious mutations (nonsense, insertion, and deletion mutations) were performed (Fig. 1). All of these variants showed no residual GALC enzyme activity above that of untransfected COS1 cells, not only validating this method but confirming their classification as severe, disease-causing mutations (Wenger et al., 2013). Infants placed in the high-risk category for KD were compound heterozygotes of mutations such as p.R111X and c.1125delTTinsGAA (p.H375Qfs*3), e.g., in the Orsini et al. (2016) study, case No. 8 (p.H375Qfs*3 + p.I546T//c.-348C>T + p.A5P + p.D232N + p.Y303C) and case No. 13 (p.R63C + p.I546T//p.R111*). However, p.R204X was identified by NBS in several infants who had polymorphisms only on the second allele and were classified in the low- or no-risk category. Expression data shown in Figure 1 strongly correlate with the high-risk category.
Several missense mutations identified in patients with KD have been classified as severe or mild, depending on the individual patient's age at diagnosis (infantile or later onset; Wenger et al., 2013). In many cases the expression data presented here (Fig. 2A,B) strongly correlate with the previous classifications. Mutations such as p.T93A, p.G95S, p.A209E, p.I234T, p.S257F, p.E258Q, p.N279I, p.D528N, and p.A563E had enzyme activity values equivalent to the mock transfections and have been associated with the infantile form of KD when a second severe allele is present (Wenger et al., 2013). Three mutations, p.T93A and p.E258Q (Fig. 2A) and p.E182K (Fig. 3A), were also chosen for this expression study because they are located at conserved residues of the mouse and human GALC active site (Deane et al., 2011). As predicted, all three showed activity values equivalent to those with the mock transfections. E182 and E258 are the proposed catalytic (proton donor and nucleophile, respectively) residues of the active site, and T93 confers substrate specificity. The p.T93A and p.E258Q mutations are considered severe alleles (Wenger et al., 2013). Expression studies of mutations classified as mild have tended to show some residual activity compared with those classified as severe, although the residual activity was much lower than in WT.
It is important to note that, for some mutations to be associated with any form of KD (e.g., p.G41S, p.A44T, p.R63H, p.G268S, p.M309V, p.R515C, p.L618S, and p.V665M), other variants must be present in cis (the haplotype effect). All these mutations are classified as mild when they are in cis with p.I546T (Fig. 2A,B). It is worth noting that there likely are exceptions to the work presented here. For example, p.W410G (Fig. 2B) has been reported in a compound heterozygous infantile Japanese patient with KD (Fu et al., 1999); however, we detected some GALC residual activity (Fig. 2B). Because the presence of polymorphisms is not mentioned in the Fu et al. article, we can only speculate that p.W410G may have another polymorphism in cis that reduced the GALC activity.
Most individuals who were found to have a VOUS identified by NBS had polymorphisms on the second allele and were classified under the no-risk category after confirmatory testing. However, when a VOUS showed very low GALC activity in expression studies (Fig. 3A,B) and another mutation was detected on the other chromosome, the confirmatory enzyme activity usually fell in the moderate- to high-risk category. Compound heterozygotes categorized as high risk (Orsini et al., 2016) include alleles p.R63C + p.I546T, p.K83E + p.I546T, and p.M101V + c.1786 + 5C>G + p.A625T. Sequence analysis of individuals with low GALC activity detected by NBS identified several variants that have significant residual GALC activity in expression studies, even in the presence of polymorphisms (e.g., p.R53Q, p.N151S, p.V320M, p.T452S, and p.A596V; Fig. 3A,B). For example, a baby homozygous for the p.V320M + p.I546T haplotype was placed in the no-risk category after confirmatory testing. We speculate that these variants are not disease causing when found either in the homozygous state or in trans with another variant. The placing of a newborn in the high-risk category by confirmatory testing does not automatically mean that he or she will develop KD at any age.
Two variants, p.T96A and p.Y303C, identified by NBS and in confirmed patients always carry the same polymorphisms (p.A5P + p.G9G + p.D232N). These alleles are worthy of special attention because they are found at high frequency in the NBS referral population. The carrier/allele frequencies for p.T96A and p.Y303C in New York State NBS are 0.276/0.149 and 0.112/0.068, respectively (Orsini et al., 2016). However, no homozygous or compound heterozygous individuals referred with p.T96A have been classified as high risk after confirmatory testing. In contrast, 28.6% of the babies classified as high risk from the NBS population carry at least one p.Y303C allele, and one high-risk infant is homozygous for p.Y303C. Figure 4 clearly shows that there is appreciable residual activity (34.8%) for the p.T96A + p.A5P + p.G9G + p.D232N haplotype but very little residual activity (3%) for the p.Y303C + p.A5P + p.G9G + p.D232N haplotype when the enzyme activities are calculated with the expression system. There are reports of some late-onset patients who are compound heterozygotes for p.Y303C and a severe mutation, but there are no individuals with confirmed KD who are homozygous for this mutation (Wenger et al., 2014). This allele has been associated only with the late-onset form of the disease.
A 53-year-old man diagnosed with KD was found to be compound heterozygous for p.D171V + p.R168C + p.I546T/p.T96A + p.A5P + p.G9G + p.D232N + p.I546T (Luzi et al., 1996). It is important to note that none of the New York State NBS KD referrals carries the p.I546T polymorphism in cis with p.T96A. This reinforces the concept of the haplotype effect as a requirement in some cases for manifestation of KD. All of the data collected to date indicate that only when the p.T96A mutation is in cis with the p.I546T polymorphism does it become a disease-causing allele, resulting in late-onset KD (Wenger et al., 2014).
This study is very helpful for evaluating moderate differences in activities for variations of haplotypes; however, the variability of cell numbers and GALC measurements poses a limitation when discriminating infantile and later-onset forms of the disease. Nevertheless, significant information may be obtained from expression of mutations and haplotypes found in patients and in NBS. A mutation with (in cis) or without another variant or polymorphism can give significantly different GALC activity values because of the haplotype effect. The relative activities will be helpful in risk assessment of referred infants.
This is the largest study showing expression of known and novel mutations and haplotypes found in the GALC gene. These studies may provide help to NBS programs and clinicians in interpreting molecular results collected in asymptomatic infants, particularly as KD NBS expands to more states.
CONFLICT OF INTEREST STATEMENT
The authors have no conflicts of interest.
ROLE OF AUTHORS
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: CAS-M, PL, DAW. Acquisition of data: PL, MN. Analysis and interpretation of data: CAS-M, PL, MN, JJO, MC, DAW. Drafting of the manuscript: CAS-M, MN, PL, DAW. Critical revision of the article for important intellectual content: CAS-M, PL, MN, JJO, MC, DAW. Statistical analysis: CAS-M, PL. Obtained funding: DAW. Administrative, technical, and material support: PL, MN. Study supervision: CAS-M, PL, DAW.