Kaposi Sarcoma-Associated Herpesvirus Sequencing in People Living With HIV in the Southern United States Reveals Subtype Diversity and Multiple Infections
*Institute at which the work was performed.
ABSTRACT
Kaposi sarcoma-associated herpesvirus (KSHV) is the causative agent of Kaposi's sarcoma and lymphoproliferative diseases collectively identified as KSHV-associated diseases (KAD). While KAD incidence has decreased across the United States, regional and population-based variability exists, with higher rates in southern states. To understand the molecular epidemiology of KSHV in this region, samples were collected from people living with HIV (PWH) with or without history of KAD. PWH, mainly men who have sex with men (MSM), were recruited from a large, urban hospital system in Dallas, Texas, in two separate studies. The studies included 220 individuals without KAD and 59 patients with KAD. Whole blood and/or oral fluids were collected and tested by qPCR. KSHV subtypes were determined from 66 of 85 individuals with detectable KSHV loads by a combination of next-generation and targeted Sanger sequencing. All major KSHV subtypes, except D, were observed including subtypes E and F. In each of three individuals, multiple KSHV genome variants were identified. This study importantly highlights KSHV subtype diversity in the southern United States, which is an area with a high KS incidence. Genome diversity and multiple infections merit epidemiological consideration, including for the future development of vaccines.
1 Introduction
The incidence of Kaposi's sarcoma associated diseases (KAD), especially Kaposi's sarcoma (KS), has decreased significantly in people living with HIV (PWH) in the United States (U.S) since the introduction of potent combination anti-retroviral therapies (ART). The decline, however, has not been uniform in all regions of the country. Notably, recent cancer registry data shows that the incidence of KS in PWH increased among Black men in the southern US which may be due to geographical, age, and racial disparities that affect access to health care [1-4]. A nation-wide survey using 2000–2013 data from the Surveillance, Epidemiology, and End Results (SEER) database showed an increase of KS diagnoses, with an associated trend of higher mortality due to KS, in Black men when compared to other ethnicities [5]. More recently, SEER data through 2021 indicates a slight decline in KS incidence in Black and Hispanic men, however the rates remain twice that observed in non-Hispanic white men. In fact, despite observed declines across the United States, the overall trend of KS incidence across all ethnicities in the United States South remains stable and above the levels seen before the AIDS epidemic (https://seer.cancer.gov/statistics-network/explorer/) (evaluated 6/28/24).
Limited molecular epidemiology studies of KSHV have been conducted in the United States since the early 2000s and the distribution of viral subtypes is essentially unknown for most communities [6-10]. It is also possible that KSHV subtypes in circulation today may differ from those previously reported in the 1990s as recently speculated [11]. The worldwide distribution of KSHV subtypes as defined by the KSHV K1 gene sequence is regional, with some subtypes predominating within certain geographic areas or within ethnic groups [6, 12]. KSHV K1 subtypes A and C are commonly reported in Europe and Asia as well as in regions that were colonized by peoples from Europe, including North America, South America, and Australia [13, 14]. Subtypes B, and A5 are more common in populations living in Africa or of African origin [7]. The less common subtypes D, E, and F were first observed in indigenous populations in the Pacific, South America, and South Africa respectively. The KSHV K15 gene is also used for subtype analysis and is considered allelic with three recognized subtypes: P, M, and N. The K15 P and M subtypes have been reported world-wide while the N has been only found in people born in Africa based upon currently available data [15, 16].
Within the population of PWH, particularly men who have sex with men (MSM), KSHV coinfection is very prevalent. The KSHV seroprevalence in a population in Dallas, Texas was recently estimated to be 68%. It was also determined that behavioral risk factors, but not race or ethnicity, were associated with KSHV seropositivity [17]. KSHV shedding in oral fluids, which is a driver of transmission, was quite high in the same study population. Taking advantage of the high KSHV loads observed in oral fluids, as well as detectable KSHV DNA in whole blood, this study was initiated to identify the KSHV K1 subtypes in this population.
2 Materials and Methods
2.1 Characteristics of Study Population
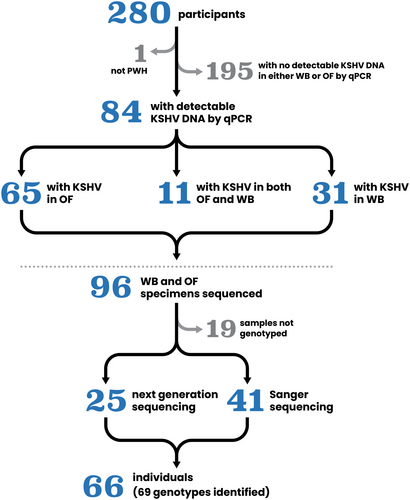
Samples from 280 individuals from two separate studies conducted at Parkland Health's outpatient HIV clinic in Dallas, Texas were sequenced. The first study was a cross-sectional cohort investigating KSHV seroprevalence which included 206 individuals with no history of KAD and 14 participants with KS, as previously reported [17]. Participants were recruited between January 2020 and September 2021. Sera were collected for serological assays while whole blood and oral fluids were collected for molecular studies.
The second study enrolled 60 PWH with history of KS, from September 2022 until April 2023. Participants provided oral fluid samples for KSHV viral load (VL) quantitation and sequencing to determine viral subtypes. Both studies were approved by the University of Texas Southwestern Institutional Review Board (STU 2019-1204 and STU 2022-0355) and all participants provided informed consent at enrollment according to the Declaration of Helsinki. Figure 1.
2.2 DNA Extraction and KSHV qPCR Assays
Whole blood was collected from study participants utilizing Qiagen PAXgene vacutainer tubes (Qiagen, Hilden, Germany), while oral fluids were collected in mouthwash. Samples were processed and DNA extracted as previously reported [17]. KSHV VL was measured in the extracted DNA using qPCR assays targeting the human endogenous retrovirus 3 gene (ERV-3), used as a cell quantitation marker, and the KSHV K6 gene region [18, 19].
2.3 Library Preparation and NGS Sequencing
Genomic DNA was fragmented using a Covaris focused-ultrasonicator (Covaris, Woburn, MA, USA). Sequence libraries were generated using two different target enrichment kits: Agilent SureSelect XT (Agilent Santa Clara, CA) [10, 20, 21] and KAPA HyperCap (KAPA Biosystems Inc, Wilmington, MA). The genomic DNA input ranged from 200 ng to 3 µg for SureSelect and 100 ng for HyperCap. Both library preparation kits contain KSHV specific bait sets based on representative KSHV genomes of all K1 and K15 subtypes.
Samples were sequenced with Illumina MiSeq and NexSeq. 2000 instruments (Illumina, Hayward, CA) with either library preparation method generating 250 bp or 150 bp paired end reads. Some samples were sequenced with both methods; four, UTSW101, UTSW113, UTSW139, and UTSW141 to resolve poor KSHV coverage due to interference from high Epstein-Barr virus (EBV) load and one, UTSW107, to confirm KSHV multiple infections observed with SureSelect XT (Supporting information Table S1).
2.4 Viral Genome Assemblies
Near full-length KSHV genomes were generated combining reference-guided alignment against the KSHV reference genome NC_009333.1 (GK18) with a de novo assembly approach [9, 10]. The internal repetitive regions (NC_009333.1:g.24230-25045, 29927-30055, 118229-113914, 124784-126456, and 137169-137969), and the large terminal repeats between the KSHV K1 and K15 genes were masked in the final alignments as they cannot be computationally resolved. Samples with read depth coverage less than 30X or with high percentages of unresolved base calls were not assembled (Supporting information Table S1). All assembled genomes were manually curated to confirm sequence variants and to resolve areas of high variability including K1, vIRF-2, and K15 gene regions. Variable gene regions were manually examined for evidence of multiple infections observed as overlapping reads with distinctive polymorphisms. Additionally, de novo K1 and K15 gene-specific subassemblies were used to distinguish KSHV subtypes by mapping overlapping reads in these highly variable regions [10]. The curated sequence alignments were exported from Geneious (Geneious Prime 2022.0.2) as consensus FASTA files for submission to GenBank and further downstream analyzes [22].
2.5 Sanger Sequencing of the K1 Gene
For samples with KSHV VL estimates determined to be suboptimal for NGS, KSHV K1 subtype was identified by Sanger sequencing of nested-PCR products as previously reported. Briefly, the outer nested primers used were ATGTTCCTGTATGTTGTCTGC (outer forward) and AGTACCAATCCACTGGTTGCG (outer reverse) followed by inner nested primers GTCTGCAGTCTGGCGGTTTGC (inner forward) and CTGGTTGCGTATAGTCTTCCG (inner reverse). The PCR cycling conditions for both rounds of PCR were similar, consisting of 1 min 45 s at 95°C and 35 cycles of 1 min at 96°C, 45 s at 51°C (outer nest) and 58°C (inner nest), and 1 min at 72°C. The inner nested procedure used 5 µl of first round product and both rounds ended with a 5-min hold at 72°C. The final K1 sequence product size was 840 base pairs [23, 24]. All Sanger sequencing was performed using an Applied Biosystems 3130XL genetic sequencer (Thermo Fisher Scientific). The sequencing of each sample was performed multiple times and a minimum of 4 overlapping reads were used to assemble the K1 gene sequences [9].
2.6 Phylogenetic Analyzes
Three samples, UTSW107, UTSW595, and UTSW601 had unresolvable genomes indicating infections with multiple KSHV genomes and were consequently excluded from phylogenetic analysis. The remaining 22 new near full-length consensus sequences were aligned with 35 published KSHV genomes featuring all available K1 and K15 gene subtypes, using MAFFT v7.511 and default settings before importation into Geneious (Geneious Prime 2022.0.2) [22]. The internal repeat regions were masked in the alignment, and the remaining nucleotide bases were used for phylogenetic analysis. A SplitsTree v4.15.1 analysis was performed using the masked alignment [25]. The Neighbor-Net method with default settings of 1000 bootstrap replicates was used to construct a phylogenetic tree [26].
KSHV K1 gene subtypes were identified using a combination of BLAST similarity searches [27] followed with confirmation by phylogenetic tree analysis. The K1 gene sequences were translated to amino acid and an alignment was made in Geneious using the MAFFT module (v1.5.0) with default settings (Geneious Prime 2022.0.2) [22]. Individual K1 sequences in four samples with multiple KSHV infections were obtained using the K1 de novo subassembly for inclusion in the final alignment. The resulting amino acid alignments, including 63 publicly available KSHV subtype-specific references, were used to infer phylogenomic trees via the neighboring-joining method using IQTree version 2.2.0.5 [28] with default settings. The IQTree output file was visualized in FigTree version v1.4.4 (https://tree.bio.ed.ac.uk/software/figtree).
2.7 Analysis of Co-Infections by Multiple KSHV Genomes
Dot plots mapping the variant nucleotide positions across the KSHV genomes for the three multiple infection samples, UTSW595, UTSW601, and UTSW107 used previously published methods [10]. Visual examples of reads constituting mixed infections were illustrated for each genome using the K1, ORF25 and ORF47 gene regions respectively, generated by the reference guided assembly pipeline, and exported as screen shots from Geneious (Geneious Prime 2022.0.2) [22].
3 Results
3.1 KSHV DNA Load Measurement
The VLs of samples collected during the 2020–2021 recruitment period were previously reported for the 206 individuals without history of KAD [17]. In the current study, 14 individuals from the earlier study and an additional 59 people with history of KAD recruited more recently were tested by qPCR. KSHV DNA was detected by qPCR in 30% (85 of 280) of study participants in either whole blood or oral fluids (Table 1).
| Individual ID | Country born | Race/ethnicity | KAD | Age | Material | HIV status | KSHV copies per million cell equivalents | KSHV subtype (K1/K15 genes) |
|---|---|---|---|---|---|---|---|---|
| UTSW500 | USA | Black | 23 | OF | Pos | 60 000 | A1/P | |
| UTSW523 | USA | Black | 41 | OF | Pos | QP | ND | |
| UTSW535 | USA | Black | 47 | OF | Pos | 19 000 | A5/P | |
| UTSW540 | USA | Black | 35 | OF | Pos | 56 250 | ND | |
| UTSW546 | USA | White | 51 | OF | Pos | 2600 | A4 | |
| UTSW547 | USA | Black | 32 | OF | Pos | 280 850 | A4/M | |
| UTSW548 | Colombia | Hispanic | 38 | OF | Pos | 3000 | A4 | |
| UTSW556 | USA | Black | KS | 39 | OF | Pos | 4750 | C7 |
| UTSW557 | El Salvador | Hispanic | 33 | OF | Pos | 126 300 | A4/P | |
| UTSW559 | USA | White | 40 | OF | Pos | 1 354 150 | A4 | |
| UTSW560 | Mexico | Hispanic | 57 | OF | Pos | 57 895 | E2/M | |
| UTSW562 | USA | Black | 57 | OF | Pos | 8000 | C3 | |
| UTSW564 | USA | Black | KS | 32 | OF | Pos | 1100 | C3 |
| UTSW567 | USA | Other | 37 | WB | Pos | QP | ND | |
| UTSW568 | Mexico | Hispanic | KS | 44 | WB | Pos | QP | C3 |
| UTSW569 | USA | Hispanic | 49 | WB | Pos | < 3 | A3 | |
| UTSW571 | USA | White | 44 | OF | Pos | 600 000 | B1/M | |
| UTSW575 | USA | Hispanic | 47 | OF | Pos | 1 314 250 | A4/P | |
| UTSW578 | Mexico | Hispanic | 54 | WB | Pos | QP | ND | |
| UTSW583 | USA | Black | 58 | OF | Pos | 215 385 | A4 | |
| UTSW586 | Mexico | Hispanic | KS | 38 | WB | Pos | 255 | C7 |
| UTSW592 | USA | Hispanic | 32 | OF | Pos | 4440 | A4 | |
| UTSW595 | USA | White | KS | 62 | OF | Pos | 1 000 000 | F2, C2/M |
| UTSW598 | USA | Black | 27 | WB | Pos | QP | ND | |
| UTSW601 | USA | Black | 36 | OF | Pos | 380 000 | C1, C7/P | |
| UTSW603 | USA | Hispanic | 31 | OF | Pos | QP | C3 | |
| UTSW604 | USA | Black | KS | 34 | OF | Pos | QP | A4 |
| UTSW605 | USA | Black | KS | 40 | OF | Pos | 91 650 | C7/P |
| UTSW607 | USA | Black | 56 | OF | Pos | 33 735 | A2/M | |
| UTSW613 | Colombia | Hispanic | 30 | WB | Pos | QP | A3 | |
| UTSW615 | USA | Hispanic | 24 | OF | Pos | 14 615 | F2 | |
| UTSW617 | USA | Black | 34 | OF | Pos | 375 | A4 | |
| UTSW620 | USA | Black | 55 | WB | Pos | 415 | ND | |
| UTSW621 | USA | Hispanic | 53 | OF | Pos | 560 000 | A2 | |
| UTSW622 | USA | Black | 51 | OF | Pos | 2 800 000 | C1 | |
| UTSW623 | USA | Asian | 57 | OF | Pos | 17 330 | C3 | |
| UTSW624 | USA | White | 70 | OF | Pos | 68 085 | F2/M | |
| UTSW627 | USA | White | 45 | OF | Pos | 1635 | C3 | |
| UTSW628 | USA | Hispanic | 37 | OF | Pos | 2025 | C3 | |
| UTSW631 | USA | Black | 31 | WB | Pos | 435 | A1 | |
| UTSW637 | USA | Black | 19 | WB | Pos | QP | ND | |
| UTSW645 | USA | Black | 32 | OF | Pos | 13 000 | A5 | |
| UTSW646 | USA | White | 59 | OF | Pos | 916 665 | C1/M | |
| UTSW651 | USA | White | 52 | WB | Pos | 680 | ND | |
| UTSW652 | Mexico | Hispanic | 61 | WB | Pos | QP | ND | |
| UTSW657 | Mexico | Hispanic | 27 | OF | Pos | 4 558 140 | E2/M | |
| UTSW662 | USA | White | 53 | OF | Pos | 46 510 | A4 | |
| UTSW666 | USA | Black | 28 | OF | Pos | 139 620 | ND | |
| UTSW668 | USA | White | 51 | OF | Pos | QP | ND | |
| UTSW670 | Honduras | Hispanic | 26 | WB | Pos | QP | ND | |
| UTSW675 | USA | Black | 23 | OF | Pos | QP | A4 | |
| UTSW676 | USA | Hispanic | 51 | OF | Pos | 6665 | C3 | |
| UTSW677 | Mexico | Hispanic | 60 | WB | Pos | 180 | A4 | |
| UTSW679 | USA | White | 55 | OF | Pos | 638 300 | C1 | |
| UTSW683 | El Salvador | Hispanic | 37 | OF | Pos | 57 140 | A5 | |
| UTSW689 | USA | White | 47 | OF | Pos | 14 440 | A1 | |
| UTSW692 | USA | Black | 35 | WB | Pos | QP | ND | |
| UTSW702 | USA | Black | 40 | OF | Pos | 23 635 | C1 | |
| UTSW704 | USA | White | 44 | OF | Pos | 7855 | A3 | |
| UTSW706 | Mexico | Hispanic | 52 | WB | Pos | QP | ND | |
| UTSW707 | USA | Hispanic | 27 | OF | Pos | 1 259 260 | A4 | |
| UTSW708 | Honduras | Hispanic | 48 | WB | Pos | QP | ND | |
| UTSW709 | USA | Black | 32 | OF | Pos | QP | A4 | |
| UTSW712 | USA | Black | 35 | OF | Pos | 475 | C3 | |
| UTSW714 | Mexico | Hispanic | 56 | WB | Pos | QP | ND | |
| UTSW717 | USA | Black | 49 | WB | Pos | QP | ND | |
| UTSW718 | Mexico | Hispanic | 38 | WB | Pos | QP | ND | |
| UTSW101 | USA | Hispanic | KS | 28 | OF | Pos | 10 910 | A1/P |
| UTSW105 | Mexico | Hispanic | KS | 43 | OF | Pos | 705 880 | A4/P |
| UTSW107 | USA | Black | KS | 45 | OF | Pos | 7825 | C3, B1/M, P |
| UTSW109 | Mexico | Hispanic | KS, MCD | 41 | OF | Pos | QP | A4 |
| UTSW111 | USA | Hispanic | KS | 32 | OF | Pos | 250 | A4 |
| UTSW113 | USA | Hispanic | KS | 43 | OF | Pos | 8380 | F2/M |
| UTSW118 | USA | White | KS | 62 | OF | Pos | 21 110 | F2/M |
| UTSW119 | USA | White | KS | 55 | OF | Pos | 947 365 | A4/P |
| UTSW124 | Democratic Republic of Congo | Black | KS | 54 | OF | Pos | 700 | B1 |
| UTSW125 | USA | Black | KS | 41 | OF | Pos | 4365 | C3 |
| UTSW130 | USA | White | KS | 64 | OF | Pos | 31 665 | F2/M |
| UTSW132 | USA | Hispanic | KS & MCD | 32 | OF | Pos | 123 635 | C3 |
| UTSW136 | Cameroon | Black | KS | 69 | OF | Neg | 4910 | B1, A4/P |
| UTSW137 | USA | Hispanic | KS | 56 | OF | Pos | 40 870 | A5/P |
| UTSW138 | Mexico | Hispanic | KS | 53 | OF | Pos | 330 | A4 |
| UTSW139 | USA | Black | KS | 42 | OF | Pos | 3220 | C7/P |
| UTSW141 | USA | Hispanic | KS | 43 | OF | Pos | 105 880 | A4/B1 |
| UTSW144 | Mexico | Hispanic | KS | 47 | OF | Pos | 49 230 | A1/M |
- Abbreviations: KS, Kaposi's sarcoma; MCD, multicentric castleman disease; OF, oral fluids; WB, whole blood; ND, not determined.
3.2 Patient Characteristics
The characteristics of individuals with detectable KSHV DNA assessed for sequencing are shown in Table 1. The participants were men living with HIV with a median age of 43 (range 19–70 years), and 38% had histories of KAD. The median HIV load in the entire cohort was 24 copies/ml with an interquartile range of 0 to 66. All participants were prescribed ART; however, adherence was not assessed in this study. A total of 22 individuals from both studies had active KS at the time of enrollment. The population was predominantly Hispanic (41%) and Black (36%) with 22% White participants. KSHV K1 A and C subtypes predominate in the study (82%).
3.3 Newly Sequenced Near Full-Length KSHV Genomes and K1 Gene Subtyping
Fifty-nine of the 85 samples with detectable KSHV DNA by qPCR were Sanger sequenced to determine K1 gene subtype, which was successful in 41 participants as summarized in Table 1 and shown in Figure 2. A wide variety of KSHV K1 subtypes were observed including A1, A2, A3, A4, A5, B1, C1, and C3, as well as less frequently observed subtypes C7, E2, and F2. Data is available in GenBank using accession numbers PP789823-PP789863; PP952419-PP952426.
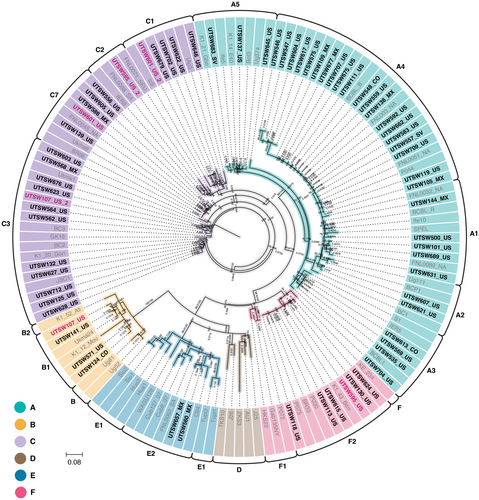
Using the KSHV load measured by qPCR, oral fluid DNA samples from twenty-six individuals were selected for NGS. Near full-length genomes were successfully obtained from 22 samples at greater than 30X (range 38X-5622X) read depth coverage (Supporting Information Table S1). One sample, UTSW124, with an estimated input of 17 KSHV copies/100 ng DNA, was sequenced by KAPA HyperPrep during optimization of the protocol to inform lower limit of KSHV load required for full genome coverage. UTSW124 was not included in full genome analysis, but the K1 gene sequence was included in the KSHV subtype analysis. The twenty-two new near full-length KSHV genomes were analyzed using SplitsTree v4.15.1 (Figure 3) and are available in GenBank as accessions PP768312-PP768333 [26].
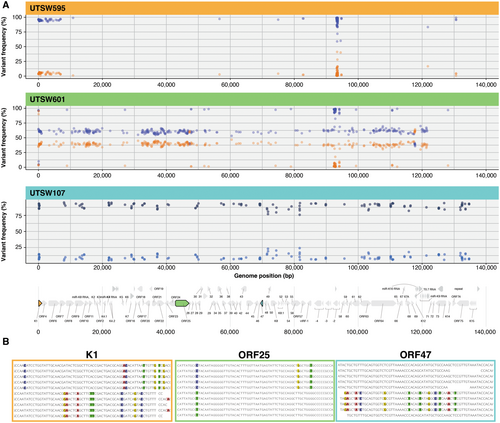
The individual KSHV genomes for samples UTSW595, UTSW601, and UTSW107 were unresolvable and had clear evidence of more than one KSHV genome. This is shown by subtype specific variations within variable gene regions spanning across the viral genome (Figure 4). Infection with multiple lineages was confirmed for UTSW107 using two independent library preparations. To visualize the mixed infections across the KSHV genome, positions of nucleotide polymorphisms identifying the mixed infections are plotted based on frequency of occurrence (Figure 4A). Minority genomes constituting less than 5% of reads are more easily detectable in highly variable gene regions like K1, as demonstrated for UTSW595, while multiple genomes with coequal frequencies are distinguishable across the viral genome, as shown in UTSW601 (Figure 4A).
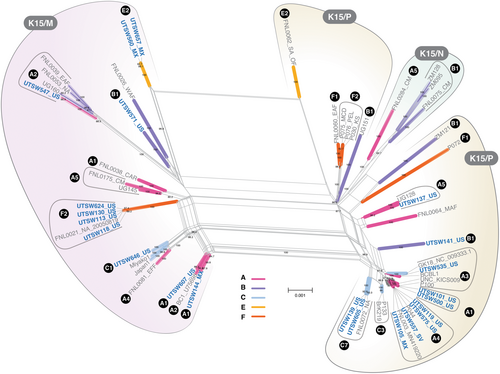
A wide range of KSHV K1 subtypes were identified in the new near full-length KSHV genomes, including the E2 and F2 for which limited information is available (Figure 3). K15 gene subtypes P and M were observed, while the N, more commonly seen in people born in Africa, was not present in the data set. Both individuals with KSHV K1 E2 subtypes were born in Mexico. Both E genomes were K15 M subtypes, in contrast the only other E genome currently available is from South America and has a K15 P allele (Figure 3). All participants with K1 F2 subtypes were born in the United States.
3.4 KSHV Gene Variations
Inspection of the genomes confirmed KSHV K1 subtype-specific indels and structural features recently described [10] including a sequence inversion between ORF8 and ORF9 observed in sequences with a F2 K1 subtype namely UTSW113, UTSW118, UTSW130 and UTSW624. The F2 subtype samples also have a two amino acid deletion in the ORF64 CDS shared with K1 A and C subtype samples UTSW535 and UTSW646. A K3 amino acid insertion was previously reported in samples of persons not born in Africa. This same K3 insertion was observed in K1 A subtype samples UTSW101, UTSW105, UTSW144, UTSW500, UTSW557, UTSW575, and UTSW607. A previously reported subtype-specific sequence inversion between ORF9-10 was observed in K1 B1 subtype sample UTSW547. Interestingly, the two K1 E2 genomes UTSW560 and UTSW657 share a distinctive 303 bp deletion between miR-K12-6 and miR-K12-5 with the only other published E2 sequence available, FNL0062 [10]. Additional observed structural features are summarized in Table 2.
| Gene/Function | Indel description | Reference position | UTSW sequences with Indel |
|---|---|---|---|
| ORF8-9 intron | 26 bp inversion | NC_009333.1:g.11277_11302inv | 113, 118, 130, 595, 624 |
| ORF9-10 intron | 80 bp inversion | NC_009333.1:g.14374_14465inv | 547 |
| ORF9-10 intron | 4 bp deletion | NC_009333,1:g.14393_14396del | 113, 118, 130, 137, 141, 547, 560, 571, 624, 657 |
| K3/E3 ubiquitin ligase; downregulation of MHC-I | 11 amino acid insertion QDGPAAGAPGN |
NC_009333.1:g.18933_18934ins GGAGCTGCCCCCGCGGGGCCATTTTGGTCGCCT |
101, 105, 144, 500, 557, 575, 607 |
| ORF45/virion phosphoprotein; inhibition of IRF-7 | Series of polymorphisms A67D, D69E, P71L, 105insD, H127N |
NC_009333.1:g.68297 G > T (H127N) NC_009333.1:g.68395_68396ins GTC (D insertion) NC_009333.1:g.68464 G > A (P71L) NC_009333.1:g.68469 G > T(D69E) NC_009333.1:g.68476 G > T (A67D) |
113, 118, 130, 141, 547, 624 |
| ORF46/viral uracil DNA glycosylase | Amino acid changes I16N, K67R, K92R, G219A, H221Y * | NC_009333.1:g,69457 T > A (I16N) NC_009333.1:g. 69336 A > G (K67R) NC_009333.1:g.69304 A > G (K92R) NC_009333.1:g.68848 G > C (G219A) |
560, 657 |
| ORF47/envelope glycoprotein gL | Series of polymorphisms T66K, G67D, D68I, W94G, T111A, T114A, A119E, D123N, 125delSIHNV, N130S, I132L |
NC_009333.1:g.69621 T > G (I132L) NC_009333.1:g.69625 G > A (N130S) NC_009333.1:g.69626 T > C (N130S) NC_009333.1:g.69628_69642del (SIHNV deletion) NC_009333.1:g.69646 A > G (D123N) NC_009333.1:g.69648 C > T (D123N) NC_009333.1:g.69659 G > T (A119E) NC_009333.1:g.69675 T > C (T114A) NC_009333.1:g.69679 G > T NC_009333.1:g.69682 C > T (T111A) NC_009333.1:g.69684 T > C (T111A) NC_009333.1:g.69688 A > G NC_009333.1:g.69691 T > C NC_009333.1:g.69735 A > C (W94G) NC_009333.1:g.69736 G > T NC_009333.1:g.69745 T > C NC_009333.1:g.69754 G > C NC_009333.1:g.69774 T > G NC_009333.1:g.69775 C > G NC_009333.1:g.69778 A > G NC_009333.1:g.69784 C > T NC_009333.1:g.69790 C > A NC_009333.1:g.69793 G > A NC_009333.1:g.69796 T > G NC_009333.1:g.69812 T > A (D68I) NC_009333.1:g.69813 C > T (D68I) NC_009333.1:g.69814 G > A (G67D) NC_009333.1:g.69815 C > T (G67D) NC_009333.1:g.69818 G > T (T66K) NC_009333.1:g.69820 T > C NC_009333.1:g.69829 C > A |
113, 118, 130, 139, 605, 624, 646 |
| ORF64/large tegument protein | Two amino acid deletion 2265delGQ |
NC_009333.1:g.110899_110904del (GQ deletion) | 113, 118, 130, 535, 624, 646 |
| ORF74/tegument protein | Amino acid deletion 12delD |
NC_009333.1:g.129553_129555del (D deletion) |
137 and 547 |
| microRNA cluster region | 303 bp deletion microRNA coding region | NC_009333.1:g.121190_121492del | 560 and 675 |
4 Discussion
The southern US, including Texas, continues to experience a comparatively higher incidence of KS compared to other US regions, particularly in Black men, despite widespread availability of anti-retroviral medications [1-3]. Our previous studies showed a high KSHV seroprevalence of 68% in MSM with HIV in Dallas, Texas [29], as well as high levels of KSHV DNA in oral fluids. Behavioral factors associated with KSHV seropositivity included self-reported use of methamphetamines and oral-anal and/or oral-penile sex [17]. Additionally, persistent racial health care disparities limit access to treatment, which affects outcomes [30]. In the current study, the distribution of KSHV subtypes was investigated in samples selected based upon the KSHV load results in whole blood and oral fluids.
While A and C subtypes were the most common, a striking variety of KSHV K1 sequences was found, including all major subtypes except D, which likely reflects the diverse underlying population in the Dallas, Texas area. The E2 subtype was observed in two individuals born in Mexico. We recently reported a similar diversity of KSHV K1 subtypes among patients enrolled in clinical studies at the HIV and AIDS Malignancy Branch (HAMB) of the National Cancer Institute in Bethesda, Maryland [10]. The populations differed between the studies, however, as people referred to the NCI for KAD were born in 22 different countries whilst in this study, participants were recruited in a community-based setting and were mostly born in the U.S. (63 of 85, 74%) or neighboring Mexico (14 of 85, 16%).
Studies of KSHV subtypes in the United States have been sparse for the last twenty years and the current study suggests that the distribution of viral subtypes in the context of high KSHV seroprevalence may be more diverse than appreciated. The area that is now the state of Texas has been settled by people of diverse ancestries over the course of the last several hundred years. A 2001 study of KSHV genotypes conducted in a population of PWH and individuals with classical KS in the San Antonio area, primarily with Hispanic ancestry, found a similar predominance of C and A K1 subtypes [31]. Notably many of the sequences obtained from Hispanic participants in the 2001 study were of the K15 M subtype [31] which was also observed in the current study. High KSHV K1 subtype diversity reported in recent studies from Spain and Ireland suggests that KSHV genomes may not be as localized to specific regions as previously assumed [32, 33]. Interestingly, a study from Brazil of 550 KSHV K1 sequences published in GenBank between the years 1997–2020 reported a similarly wide subtype distribution [14]. In contrast, in a KS case-control study conducted in Cameroon we observed only A5 and B K1 subtypes [21]. The current KSHV genome worldwide distribution patterns rely upon available sequencing data which is disproportionally biased to genomes obtained from samples with higher KSHV viral copies. Sometimes referred to as convenience samples, they have done much to inform but do not necessarily reflect the true viral genome distribution as not all members of any specific population are equally represented. Recent improvements in sequencing technologies, including whole genome next-generation platforms, allow the sequencing of samples with lower KSHV estimated VL. It is possible that with increased KSHV genome sequencing efforts, a more informative distribution of viral subtypes within regions of high associated disease incidence can be obtained.
In three individuals, one of whom, UTSW601, did not have KS disease, multiple KSHV infections were identified by NGS. This is the latest of several recent reports of individuals with more than one detectable KSHV genome, adding strength to the hypothesis that multiple infections may not be uncommon [9, 10, 21]. Accumulating evidence of infections by multiple genomes suggests that pre-existing KSHV infections may not be protective of infection against infection during subsequent exposures. Whether multiple infections occur simultaneously or are acquired sequentially during the lifetime of an individual cannot be determined in these cross-sectional data. The observation of multiple KSHV genomes in oral fluids has implications for the understanding of viral transmission and the development of a KSHV vaccine for which efforts are intensifying [34-37]. The new KSHV genomes were also examined for structural variations and compared to publicly available sequences as summarized in Table 2. Many indels previously reported are also observed in this study, including sequence inversions and large deletions [10]. Polymorphisms in the ORF46 gene encoding viral uracil DNA glycosylase, which have previously been shown to affect the function of the protein, were present in the current E2 study genomes, except for H221Y, the second variation noted within the extended leucine loop. The consequences of the specific series of variations observed in the new E2 sequences (I16N, K67R, K92R, and G219A) in terms of the function of the viral uracil DNA glycosylase are currently unknown [10, 38].
In summary, the results of this study of KSHV K1 subtype distribution in a single institution in the Dallas, Texas area show an unexpectedly high diversity of viral genomes. The observed wide variety of subtypes likely reflects the rich and long-standing ethnic diversity in the region and the mobility of its population. It could also reflect high KSHV transmission in highly populated cities in general. Other communities with diverse populations may harbor KSHV K1 subtypes not commonly reported in North America, which has implications for viral dynamics and evolution. The near-full length genomes obtained in this study informs global KSHV genetics and facilitates efforts to characterize viral genomes outside of the variable K1 and K15 gene regions. Importantly, analysis of the twenty-two new genomes confirms previous observations of variations possibly associated with KSHV K1 subtypes, which may ultimately allow a new definition of KSHV specific genotypes using whole viral genomes. As additional KSHV genomes become available, variations can be more precisely analyzed to determine any contribution of sequence variations to KSHV disease, transmission, and outcomes. Future efforts to more precisely define circulating KSHV subtypes within populations can benefit from advantages in sequencing techniques that allow samples of lower estimated viral load to be evaluated, thereby expanding surveys to include more of the general population without disease.
Author Contributions
Authors Vickie A. Marshall, Sheena M. Knights, Nazzarena Labo, Elizabeth Y. Chiao, Susana M. Lazarte, Elizabeth Y. Chiao, Denise Whitby, and Ank E. Nijhawan designed the study. Sheena M. Knights, Susana M. Lazarte, and Ank E. Nijhawan oversaw recruitment, provided participant care, and supervised protocols. Vickie A. Marshall, Nazzarena Labo, Wendell J. Miley, Isabella Liu, Charles A. Goodman, and Elizabeth Y. Chiao curated data including bioinformatics and statistical analyzes. Vickie A. Marshall and Kyle N. Moore preformed qPCR testing and Wendell J. Miley provided serological assay support. Vickie A. Marshall, Isabella Liu, and Elizabeth Y. Chiao conducted all sequencing protocols. Brandon F. Keele and Christine M. Fennessey provided technical support for whole genome sequencing applications. All authors contributed to manuscript writing, review, and editing.
Acknowledgments
We would like to thank the participants and their families without whom this study would not have been possible. Special thanks also to Leslie Lipkey and Agatha Macairan from the Retroviral Evolution Section, ACVP for technical support and Graphics designer Joseph Meyer at Scientific Publications, Graphics, and Media, FNLCR for his expertise in figure conceptualization and design. This study was supported by the National Center for Advancing Translational Science (NCATS) [grant number 1UL1TR003163-02] (SK, AN), as well as a Translational Pilot Program Award from the Simmons Comprehensive Cancer Center at the University of Texas Southwestern Medical Center (SK, AN). Additionally, this project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. 75N91019D00024/HHSN261200800001E (VAM, NL, IL, WJM, EMC, CAG, CMF, KNM, BFK, and DW). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Ethics Statement
Studies were approved by the University of Texas Southwestern Institutional Review Board (STU 2019-1204 and STU 2022-0355) and all participants provided informed consent at enrollment according to the Declaration of Helsinki.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. KSHV sequence information is available in GenBank referencing accession numbers PP768312-PP768333 and PP789823-PP789863; PP952419-PP952426.




