Performance evaluation of the MAGLUMI Hepatitis B virus surface antigen chemiluminescence immunoassay
Lihong Shen, Yun Zhang, and Min Shi contributed equally to this work and share first authorship.
Abstract
A highly sensitive and reliable Hepatitis B virus surface antigen (HBsAg) measurement is essential to universal screening, timely diagnosis, and management of Hepatitis B virus (HBV) infection. This study aimed to evaluate the performance of MAGLUMI HBsAg chemiluminescence immunoassay (CLIA). MAGLUMI HBsAg (CLIA) was compared against ARCHITECT HBsAg. 411 HBsAg positive samples, including different stages of infection, genotypes, subtypes, mutants, and 30 seroconversion panels were tested to evaluate diagnostic sensitivity. Diagnostic specificity was evaluated by testing 205 hospitalized samples and 5101 blood donor samples. Precision, limit of blank (LoB), limit of detection (LoD), and linearity were also verified. The diagnostic sensitivity of the MAGLUMI HBsAg (CLIA) was 100% with better seroconversion sensitivity than ARCHITECT HBsAg. The MAGLUMI HBsAg (CLIA) has optimal detection efficacy for HBV subgenotypes samples. The analytical sensitivity is 0.039 IU/mL. The initial diagnostic specificity is 99.63% on blood donors and 96.59% on hospitalized samples. The verification data demonstrated high repeatability, a LoB of 0.02 IU/mL, LoD of 0.05 IU/mL and an excellent linearity of 0.050–250 IU/mL (R2 = 0.9946). The MAGLUMI HBsAg (CLIA) is proved a highly sensitive and reliable assay with optimal subgenotype detection efficacy.
1 INTRODUCTION
Hepatitis B, spreading through blood, vaginal fluid, and semen, is one of the most common liver infections globally and the leading cause of liver fibrosis, liver cirrhosis, and liver cancer.1 It has been reported to result in 296 million people infected and 820 000 deaths in 2022, with incidence rates increasing slightly by 0.51% per year.2 Although the World Health Organization has set the goal of eliminating viral hepatitis by 2030, the annual global deaths from Hepatitis B virus (HBV) and HBV-related liver complications are expected to increase by 39% from 2015 to 2030 because of under-diagnosis and under-treatment.2 Research has shown that from 2013 to 2018, approximately 65.6% of US adults with chronic HBV infection were unaware of their HBV infection, emphasizing the importance of universal screening and timely diagnosis of HBV infection.3, 4 According to the data from WHO regional and country offices, the percentage of HBV infections undiagnosed is 89.7% globally, indicating that the global diagnosis coverage is still low and new global strategies for 2022–2030 are recommended to scale up HBV screening for the improvement of diagnosis coverage, especially in low-income and middle-income countries.5, 6 Moreover, based on the findings of a modeling study, more than 90% of diagnoses of people living with HBV infection and 80% of testing of eligible patients are necessary to reach the 2030 HBV elimination goal.6 In favor of this goal, the Centers for Disease Control and Prevention recommends that every adult aged no less than 18 years should screen for HBV infection at least once during a lifetime using three laboratory tests, including Hepatitis B surface antigen (HBsAg), antibody to hepatitis B surface antigen (anti-HBs), and total antibody to hepatitis B core antigen (anti-HBc), to help in determining HBV infection status.7, 8 Therefore, the development of fast, highly sensitive and accurate methods of HBsAg detection would contribute to the universal screening and diagnosis of HBV infection.
HBsAg, circulating in blood of HBV-infected patients, is the determinant of the large, middle, and small envelop proteins which are key components for the entry of HBV to hepatocytes.8 Meanwhile, based on different HBsAg serological reactivities, HBV can be categorized into four serotypes including adw, adr, ayw, and ayr.9 HBsAg has been discovered with different ratios of expression and secretion among all 10 HBV genotypes (A–J) and numerous subgenotypes which is characterized by genomic diversification.9 The presence of HBsAg in blood typically indicates the presence of HBV and plays a crucial role in the diagnosis of both acute and different stages of chronic hepatitis B infection as well as the antiviral therapy selection and monitoring.9 Specifically, the positive detection results of HBsAg and IgM antibody to the hepatitis B core antigen (IgM anti-HBc) within 3–6 months may indicate acute HBV infection.10, 11 If the presence of HBsAg and total anti-HBc persists for more than 6 months, the patients would be diagnosed with chronic hepatitis B infection.12, 13 The phases of chronic hepatitis B infection could be further classified into four phases according to the quantitative levels of HBsAg, HBV DNA, and other HBV-related markers reflecting virus-host interaction, which is beneficial to the management of HBV carriers.14, 15
The detection of HBsAg is essential not only to the diagnosis and classification of both acute and different stages of chronic hepatitis B infection, but also to the monitoring and management of HBV infection. Researchers have found that the prevalence of the coexistence of HBsAg and anti-HBs in chronic HBV infectious patients was 2.8%–3.6% globally and genotype C group might have the highest prevalence among all HBV genotypes, which is mainly due to the increased immune escape mutants resulting from the rise in vaccination coverage and the application of various antiviral medications.10, 11 The coexistence has been shown to be significantly correlated with progressive liver diseases, especially severe liver fibrosis, cirrhosis and hepatocellular carcinoma.16-18 Therefore, the detection of coexistent HBsAg and anti-HBs among chronic HBV infection would be useful for timely monitoring and control of increased risks of adverse clinical outcomes.17 Furthermore, quantitative HBsAg assay used with HBV DNA has been proven to be effective for monitoring prognosis and treatment response in chronic hepatitis B infection.19 Evidence has demonstrated that HBsAg concentration of less than 1000 IU/mL would promote the accurate identification of true inactive carriers and prediction of functional cure of chronic hepatitis B.19 At week 12 of peginterferon therapy, the decreased results of quantitative HBsAg and HBV DNA are the indicators of stopping treatment in genotype D HBV-infected patients.19 Moreover, quantitative HBsAg monitoring should be carried out in patients treated with nucleos(t)ide analogs for chronic hepatitis B monthly from 6 to 12 months to predict the risk of relapse and time to stop treatment.20
To improve current clinical status in terms of universal screening and timely diagnosis of different HBV genotypes, prediction of progressive liver diseases, and monitoring prognosis and therapeutic response, a highly sensitive, accurate, and fast MAGLUMI HBsAg chemiluminescence immunoassay (CLIA) has been developed for the quantitative assessment of HBsAg in human serum and plasma. The objective of this study was to evaluate the clinical performance of MAGLUMI HBsAg (CLIA) and demonstrate the detection efficiency in different HBsAg mutants and subgenotypes by comparing it with conventional HBsAg assays. Additionally, the analytical performance in terms of intraassay precision, limit of blank (LoB), limit of detection (LoD), and linearity was further verified based on the Clinical and Laboratory Standards Institute (CLSI) documents.
2 MATERIALS AND METHODS
2.1 MAGLUMI HBsAg (chemiluminescence immunoassay [CLIA]) and reference tests
The MAGLUMI HBsAg (CLIA) is a sandwich chemiluminescence immunoassay in which HBsAg forms a sandwich between anti-HBs monoclonal antibody solid phase and another anti-HBs polyclonal antibody labeled with N-(4-Aminobutyl)-N-ethylisoluminol (ABEI). After the Starter 1 + 2 are added to initiate the chemiluminescent reaction, the light signal is measured by a photomultiplier as relative light units, which is proportional to the concentration of HBsAg present in the sample. All samples in this study were detected with the MAGLUMI HBsAg (CLIA) according to the procedure recommended in the manufacturer's instruction, including mixing the completely thawed samples to become visibly homogeneous, inspecting and assuring all samples are free of foam, fibrin, red blood cells or other particulate matter.
The reference test was used to compare and evaluate the detection results. The HBV-positive samples and seroconversion panels were detected with the Abbott CE marked HBsAg quantitative assay, ARCHITECT HBsAg (REF. 6C36). ARCHITECT HBsAg was also utilized to test the hospitalized samples and blood donor samples. The ARCHITECT HBsAg Qualitative II Confirmatory v1.0 (REF.9C94) was used to further confirm the repeatedly reactive samples during the evaluation of clinical diagnostic sensitivity and diagnostic specificity.
2.2 Seroconversion sensitivity
Seroconversion sensitivity was evaluated by comparing the number of reactive bleeds and the first positive day of each sample from 30 commercially available seroconversion panels with the MAGLUMI HBsAg (CLIA) and the ARCHITECT HBsAg.
2.3 HBV subgenotype detection
The 1st International Reference Panel for HBV genotypes for HBsAg assays (Paul-Ehrlich Institut, PEI) 6100/09 consists of 15 different panel members with HBsAg concentration of approximately 100 IU/mL determined by using the arithmetic mean IU/mL values of the CLIA HBsAg concentrations (ARCHITECT HBsAg REF. 6C36). And the 15 panel members represent the following genotypes with different origins: genotype A1_1 (South Africa), genotype A1_2 (Brazil), genotype A2_3 (Germany), genotype B1_4 (Japan), genotype B2_5 (Japan), genotype C2_6 (Japan), genotype C2_7 (Japan), genotype C2_8 (Russia), genotype D1_9 (Germany), genotype D2_10 (Russia), genotype D3_11 (South Africa), genotype E_12 (West Africa), genotype F2_13 (Brazil), genotype F2_14 (Brazil), genotype H_15 (Germany). Each vial of the panel members was prepared twofold dilution series (10 dilutions) in normal human serum negative for HBsAg and anti-HBs per the manufacturer's instructions by three independent preparations.
2.4 Analytical sensitivity determination
The 3rd International WHO standard for HBsAg (HBV genotype B4, HBsAg subtypes ayw1/adw2) (NIBSC code 12/226) was used according to the manufacturer's instructions. Twofold serial dilutions (10 dilutions ranging from 0 to 0.500 IU/mL) were made in HBsAg and anti-HBs negative human serum by three independent preparations. The analytical sensitivity, defined as the theoretical (assigned) concentration corresponding to the assay's cut-off value of 0.05 IU/mL, was calculated by linear regression analysis using the mean of triplicate determinations.
2.5 Diagnostic specificity
A total of 205 hospitalized samples and 5101 blood donor samples were detected with the MAGLUMI HBsAg (CLIA) and ARCHITECT HBsAg. All samples were detected in a single determination with each assay. Samples assessed with initially reactive results were retested in duplicate and those found to be repeatedly reactive were confirmed by the ARCHITECT HBsAg Qualitative II Confirmatory v1.0.
2.6 Analytical interference
100 potentially interfering samples collected from dialysis patients, influenza vaccine recipients, multipara pregnant women or pregnant women or positive for Anti-CMV, Anti-EBV, Anti-HAV, Anti-HCV, Anti-HEV, Anti-HIV, Anti-HSV1/2, Anti-Syphilis, Anti-VZV, c-ANCA, HAMA, Hyper IgG/IgM, Hyper IgM and rheumatoid factor were tested to evaluate the cross-reactivity performance of the MAGLUMI HBsAg.
2.7 Analytical performance verification
Intraassay precision of the MAGLUMI HBsAg (CLIA) was verified within a laboratory according to CLSI EP15-A3. Three commercial quality control materials with low, medium, and high measurand concentrations were tested in 5 days, one run per day, five replicates per run. LoB and LoD of the MAGLUMI HBsAg (CLIA) were verified according to CLSI EP17-A2. In terms of LoB, two blank samples were measured in replicates of four each day for 3 days. LoD was verified using two samples containing low amounts of measurand and then measuring in four replicates on each of the same three testing days.
Linearity was verified according to CLSI EP06-A. Six samples supplied by the manufacturer with assigned concentrations were tested in three replicates within a single run.
2.8 Statistical analysis
Excel 2019 (Microsoft Inc.) was used to calculate average value, standard deviation, coefficient of variation, proportions, and corresponding Wilson 95% confidence intervals. GraphPad Prism 9.0 (GraphPad Software) was used to determine the strength of the linear relationship between the detection values and relative assigned concentrations. A p < 0.05 is considered statistically significant.
2.9 Ethics
The clinical performance evaluation study was performed by a third-party organization (laboratories of Biomex GmbH) and the assay performance verification study was conducted by the clinical laboratory at Affiliated Jinhua Hospital, Zhejiang University School of Medicine in accordance with the guidelines of the Declaration of Helsinki and in adherence with the principles of Good Clinical Practice. All samples used in this study were residual samples obtaining a general authorization for ethical approval and signed broad consent.
3 RESULTS
3.1 Clinical diagnostic sensitivity
The diagnostic sensitivity of the MAGLUMI HBsAg (CLIA) was evaluated by testing 411 HBsAg positive samples with different stages of infection (acute/chronic), different genotypes, subtypes, and native mutants and 4 recombinant mutant samples from the Diasorin recHBsAg Mutants Panel (REF: 310259). Table 1 shows that all 411 HBsAg positive samples from different genotypes (A–H) with known classification information of subtypes (Table 2) and native mutations (Table 3) were tested as reactive. And it demonstrated that the MAGLUMI HBsAg (CLIA) has a high diagnostic sensitivity of 100% (95%CI: 99.07%–100.00%). In addition, the MAGLUMI HBsAg (CLIA) is able to detect all four most common recombinant mutant samples, offering support for the clinical performance of high diagnostic sensitivity. Thus, the MAGLUMI HBsAg (CLIA) has the capacity to detect HBsAg-positive samples with different clinical stages, genotypes, serotypes, and native or recombinant mutations.
| Genotype | Number | Reactive | Nonreactive | ||
|---|---|---|---|---|---|
| HBsAg positive samples | Unspecified genotype | 96 | 96 | 0 | |
| Genotype A | Unspecified | 25 | 25 | 0 | |
| A1 | 16 | 16 | 0 | ||
| A2 | 41 | 41 | 0 | ||
| A3 | 14 | 14 | 0 | ||
| Genotype B | Unspecified | 2 | 2 | 0 | |
| B2 | 6 | 6 | 0 | ||
| B3 | 1 | 1 | 0 | ||
| B4 | 7 | 7 | 0 | ||
| B5 | 2 | 2 | 0 | ||
| Genotype C | Unspecified | 5 | 5 | 0 | |
| C1 | 7 | 7 | 0 | ||
| C2 | 7 | 7 | 0 | ||
| Genotype D | Unspecified | 59 | 59 | 0 | |
| D1 | 21 | 21 | 0 | ||
| D2 | 18 | 18 | 0 | ||
| D3 | 21 | 21 | 0 | ||
| D4 | 3 | 3 | 0 | ||
| D7 | 2 | 2 | 0 | ||
| Genotype E | Unspecified | 47 | 47 | 0 | |
| Genotype F | Unspecified | 1 | 1 | 0 | |
| F1 | 1 | 1 | 0 | ||
| F2 | 1 | 1 | 0 | ||
| Genotype G | Unspecified | 3 | 3 | 0 | |
| Genotype H | Unspecified | 5 | 5 | 0 | |
| Total | 411 | 411 | 0 | ||
| Sensitivity | 100% | ||||
| 95%CI | 99.07%–100.00% | ||||
- Abbreviations: CLIA, chemiluminescence immunoassay; HBsAg, Hepatitis B virus surface antigen; HBV, Hepatitis B virus.
| Subtype | Number of positive samples | |
|---|---|---|
| HBsAg positive samples | adr | 19 |
| adw | 76 | |
| ay | 212 | |
| ayr | 5 | |
| ayw | 193 | |
| Unspecified | 104 | |
| Total | 411 | |
- Abbreviations: HBsAg, Hepatitis B virus surface antigen; HBV, Hepatitis B virus.
| Positive mutant samples | Number | |
|---|---|---|
| N/A | 341 | |
| Native mutant samples | D114E | 4 |
| F134L | 4 | |
| G130N | 1 | |
| G130NM133I | 1 | |
| G130NY134N | 1 | |
| G130R | 1 | |
| G130R S132Y | 2 | |
| G145A | 1 | |
| Insertion116T | 1 | |
| L209V | 4 | |
| L45T | 1 | |
| M133I | 2 | |
| M133l | 3 | |
| M133LF134L | 1 | |
| M133T | 3 | |
| M133TF134L | 1 | |
| M133TT116N | 1 | |
| P120S | 1 | |
| P120T | 2 | |
| Q129H | 1 | |
| Q129R | 1 | |
| Q129R, G130N | 1 | |
| S143L | 2 | |
| S143M | 1 | |
| T116N | 4 | |
| T118A | 7 | |
| T118A | 7 | |
| T123N | 1 | |
| T126A | 2 | |
| T126N | 1 | |
| T1312l | 1 | |
| T131lY134NS143L | 1 | |
| T143M | 1 | |
| Y134F | 2 | |
| Y134N | 1 | |
| Y137F | 1 | |
| Recombinant mutant samples | G145R | 1 |
| D144A | 1 | |
| P142L–G145R | 1 | |
| P142S–G145R | 1 | |
| Total | 40 | 415 |
- Abbreviation: HBV, Hepatitis B virus.
3.2 Seroconversion diagnostic sensitivity
HBV seroconversion sensitivity was evaluated on 30 commercially available seroconversion panels by MAGLUMI HBsAg (CLIA) and the reference test ARCHITECT HBsAg. As is shown in Table 4, the MAGLUMI HBsAg (CLIA) was able to detect more positive members in seven panels, the same members in 20 panels and fewer samples in three panels. Among these three panels, the reference test detected one or two more samples than the MAGLUMI HBsAg (CLIA). Noted that the MAGLUMI HBsAg (CLIA) detected one bleed earlier positive and turned one bleed earlier back to negative compared to the reference test during the seroconversion of SCP-HBV-004 panel (data not shown), which suggests the high seroconversion sensitivity of MAGLUMI HBsAg (CLIA). Overall, the MAGLUMI HBsAg (CLIA) was able to detect slightly more seroconversion panel members (137 of 373) than the reference test (134 of 373). Furthermore, the first positive days since a positive result of the first bleed were detected for each longitudinal seroconversion panel to assess the shortness of the early window period during the acute phase of HBV infection (Table 5). Compared to the reference test, the MAGLUMI HBsAg (CLIA) had delayed detection in three panels, the same seroconversion day in 19 panels and earlier detection in eight panels. The MAGLUMI HBsAg (CLIA) (1279 days) showed a shorter window period and a relatively higher seroconversion sensitivity than the reference test (1286 days), which suggests that the MAGLUMI HBsAg (CLIA) seems likely to reduce HBsAg window period and detect HBsAg a little earlier from the first day of HBsAg exposure comparing to ARCHITECT HBsAg.
| Panel ID | Panel members | MAGLUMI HBsAg (CLIA) | ARCHITECT HBsAg |
|---|---|---|---|
| Zeptometrix HBV6271 | 5 | 3 | 3 |
| Zeptometrix HBV6273 | 6 | 2 | 2 |
| Zeptometrix HBV6274 | 7 | 6 | 7 |
| Zeptometrix HBV6275 | 7 | 3 | 3 |
| Zeptometrix HBV6276 | 8 | 2 | 2 |
| Zeptometrix HBV6277 | 11 | 5 | 5 |
| Zeptometrix HBV6279 | 7 | 2 | 2 |
| Zeptometrix HBV6283 | 11 | 4 | 4 |
| Zeptometrix HBV6290 | 12 | 6 | 6 |
| Zeptometrix HBV9074 | 20 | 3 | 3 |
| Zeptometrix HBV11000 | 9 | 1 | 3 |
| Zeptometrix HBV11001 | 8 | 4 | 4 |
| Zeptometrix HBV11002 | 6 | 5 | 4 |
| Zeptometrix HBV11003 | 8 | 3 | 3 |
| Zeptometrix HBV11006 | 17 | 6 | 5 |
| Zeptometrix HBV11007 | 14 | 5 | 4 |
| Zeptometrix HBV11008 | 18 | 5 | 4 |
| Zeptometrix HBV11009 | 23 | 5 | 5 |
| Zeptometrix HBV11012 | 6 | 3 | 3 |
| Zeptometrix HBV11013 | 35 | 7 | 6 |
| Zeptometrix HBV11016 | 9 | 4 | 5 |
| Zeptometrix HBV11017 | 14 | 6 | 5 |
| Zeptometrix HBV11024 | 14 | 4 | 4 |
| Zeptometrix HBV11026 | 16 | 5 | 4 |
| Zeptometrix HBV11027 | 13 | 5 | 5 |
| Zeptometrix HBV11029 | 13 | 4 | 4 |
| Zeptometrix HBV11031 | 16 | 6 | 6 |
| Seracare PHM939 | 5 | 3 | 3 |
| Seracare PHM937 | 5 | 3 | 3 |
| Biomex SCP-HBV-004 | 30 | 17 | 17 |
| Total panel members detected | 373 | 137 | 134 |
- Abbreviations: CLIA, chemiluminescence immunoassay; HBsAg, Hepatitis B virus surface antigen; HBV, Hepatitis B virus.
| Panel ID | First positive day | ||
|---|---|---|---|
| MAGLUMI HBsAg (CLIA) | ARCHITECT HBsAg | Difference between two testsa | |
| Zeptometrix HBV6271 | 7 | 7 | 0 |
| Zeptometrix HBV6273 | 25 | 25 | 0 |
| Zeptometrix HBV6274 | 4 | 0 | 4 |
| Zeptometrix HBV6275 | 22 | 22 | 0 |
| Zeptometrix HBV6276 | 27 | 27 | 0 |
| Zeptometrix HBV6277 | 35 | 35 | 0 |
| Zeptometrix HBV6279 | 26 | 26 | 0 |
| Zeptometrix HBV6283 | 29 | 29 | 0 |
| Zeptometrix HBV6290 | 21 | 21 | 0 |
| Zeptometrix HBV9074 | 73 | 73 | 0 |
| Zeptometrix HBV11000 | 33 | 26 | 7 |
| Zeptometrix HBV11001 | 44 | 44 | 0 |
| Zeptometrix HBV11002 | 2 | 7 | −5 |
| Zeptometrix HBV11003 | 142 | 142 | 0 |
| Zeptometrix HBV11006 | 42 | 44 | −2 |
| Zeptometrix HBV11007 | 34 | 36 | −2 |
| Zeptometrix HBV11008 | 69 | 72 | −3 |
| Zeptometrix HBV11009 | 81 | 81 | 0 |
| Zeptometrix HBV11012 | 18 | 18 | 0 |
| Zeptometrix HBV11013 | 246 | 251 | −5 |
| Zeptometrix HBV11016 | 27 | 18 | 9 |
| Zeptometrix HBV11017 | 40 | 42 | −2 |
| Zeptometrix HBV11024 | 40 | 40 | 0 |
| Zeptometrix HBV11026 | 39 | 43 | −4 |
| Zeptometrix HBV11027 | 33 | 33 | 0 |
| Zeptometrix HBV11029 | 43 | 43 | 0 |
| Zeptometrix HBV11031 | 39 | 39 | 0 |
| Seracare PHM939 | 11 | 11 | 0 |
| Seracare PHM937 | 9 | 9 | 0 |
| Biomex SCP-HBV-004 | 18 | 22 | −4 |
| Total delayed days on 30 panels | 1279 | 1286 | −7 |
- Abbreviations: CLIA, chemiluminescence immunoassay; HBsAg, Hepatitis B virus surface antigen; HBV, Hepatitis B virus.
- a Difference between two tests <0 is the number of days reduced from HBsAg exposure to detection by MAGLUMI compared to ARCHITECT HBsAg, Difference between two tests >0 is the number of days delayed from HBsAg exposure to detection by MAGLUMI compared to ARCHITECT HBsAg.
3.3 HBV subgenotype detection efficacy
1st International reference panel for HBV genotypes for HBsAg assays (PEI 6100/09) was utilized to assess the detection efficacy of the MAGLUMI HBsAg (CLIA) to detect different HBV subgenotypes. Figure 1 illustrates the quantification differences among MAGLUMI HBsAg (CLIA) and other quantitative results obtained from six different laboratories utilizing two different quantitative HBsAg assay methods, including ARCHITECT HBsAg and HISCL HBsAg Assay Kit.20 The quantitative assays HISCL HBsAg Assay Kit (19N) and MAGLUMI HBsAg (CLIA) (MAGLUMI) showed higher values for the samples 1–15 compared to the results with the ARCHITECT HBsAg. Because these sample HBsAg concentrations are all predetermined around 100 IU/mL by ARCHITECT HBsAg,20 the higher HBsAg subgenotypes detection values by both HISCL HBsAg Assay Kit (19 N) and MAGLUMI HBsAg (CLIA) (MAGLUMI) reveal a higher sensitivity than the ARCHITECT HBsAg.
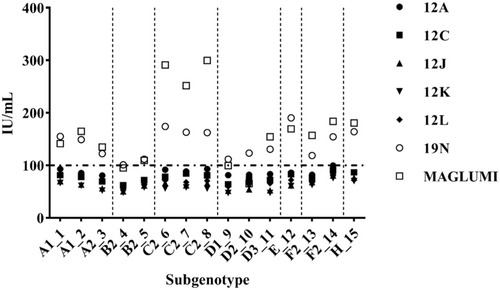
As shown in Figure 1, the MAGLUMI HBsAg (CLIA) detected subgenotype C2 with the highest concentrations among all quantitative assay results, whereas the MAGLUMI HBsAg (CLIA) generated similar values for subgenotype D2 which still showed 20.17% higher mean detection potency than the result generated by 12J at 54.6 IU/mL. Overall, the results demonstrated that except for subgenotype D2 samples, the MAGLUMI HBsAg (CLIA) successfully detected all HBV subgenotypes with higher concentrations among all quantitative tests evaluated, with an average 1.2-fold (subgenotype D2) to 5.4-fold (subgenotype C2) higher sensitivity than ARCHITECT HBsAg. Therefore, the MAGLUMI HBsAg (CLIA) has higher detection efficacy for different HBV subgenotype samples than ARCHITECT HBsAg.
3.4 Analytical sensitivity determination
To evaluate analytical sensitivity, twofold serial dilutions (10 dilutions ranging from 0 to 0.500 IU/mL) of the 3rd International WHO standard for HBsAg subtype ayw1/adw2 genotype B4 (NIBSC code 12/226) were prepared and determined by three independent preparations. The lowest concentration of the International Standard still reactive with the MAGLUMI HBsAg (CLIA) is 0.039 IU/mL, which proved that the analytical sensitivity of the MAGLUMI HBsAg (CLIA) fulfilled the criteria for the analytical sensitivity of less than 0.130 IU/mL defined by the COMMISSION IMPLEMENTING REGULATION (EU) 2022/1107 for in vitro diagnostic medical devices.
3.5 Diagnostic specificity
205 hospitalized samples and 5101 blood donor samples were detected by MAGLUMI HBsAg (CLIA) and the ARCHITECT HBsAg to evaluate the diagnostic specificity. The results presented in Table 6 demonstrate the total diagnostic specificity of the MAGLUMI HBsAg (CLIA) is 99.91% while that of the comparator assay is 99.98%. The samples initially reactive among blood donors (IR = 19) and hospitalized samples (IR = 7) were inspected and contained fibrin, which may be the results of nonspecific reaction. After centrifuging and transferring every clarified sample to a second tube for retesting, the MAGLUMI HBsAg (CLIA) gave consistent results (Supporting Information S1: Table S1), emphasizing on the correlation between sample condition, necessary preparation for analysis, and reliable results. All in all, the data in Table 6 indicates that the relatively high sensitivity performance of the MAGLUMI HBsAg (CLIA) does not compromise diagnostic specificity.
| Samples | MAGLUMI HBsAg (CLIA) | ARCHITECT HBsAg | ||
|---|---|---|---|---|
| Initial specificity | Repeat specificity | Initial specificity | Repeat specificity | |
| Blood donors (n = 5101) | 99.63% (IR = 19) | 99.92% (RR = 4) | 99.98% (IR = 1) | 99.98% (RR = 1) |
| Hospitalized samples (n = 205) | 96.59%(IR = 7) | 99.51% (RR = 1) | 100% (IR = 0) | 100% (RR = 0) |
| All negative samples (n = 5306) | 99.51% (IR = 26) | 99.91% (RR = 5) | 99.98% (IR = 1) | 99.98% (RR = 1) |
- Abbreviations: CLIA, chemiluminescence immunoassay; HBsAg, Hepatitis B virus surface antigen; HBV, Hepatitis B virus; IR, initial reactive; RR, repeat reactive.
3.6 Analytical interference
In total 100 potentially interfering samples were detected to assess the analytical specificity, and none of the samples were reactive with the MAGLUMI HBsAg (CLIA). The results are presented in Supporting Information S1: Table S2.
3.7 Analytical performance verification
As is shown in Table 7, the coefficient of variations (CVs) of the low, medium, and high samples' concentration were less than the 10% repeatability claimed by the manufacturer, which demonstrated that the repeatability of the MAGLUMI HBsAg (CLIA) is consistent with the manufacturer's claims in the instruction.
| Materials | Mean (IU/mL) | Standard deviation (IU/mL) | Coefficient of variation |
|---|---|---|---|
| Low | 0.213 | 0.008 | 3.54% |
| Medium | 122.440 | 4.718 | 3.85% |
| High | 224.120 | 6.260 | 2.79% |
- Abbreviations: CLIA, chemiluminescence immunoassay; HBsAg, Hepatitis B virus surface antigen.
Comparison of the blank results with the manufacturer's LoB claim of 0.02 IU/mL showed that all 24 measurements were equal to the claim (Supporting Information S1: Table S3). Because the observed percentage (24/24, 100%) is no less than the minimum percentage of 87%, the manufacturer's LoB claim was verified. In addition, the manufacturer's LoD claim was verified with the proportion of positive measurements between the claimed LoB (0.02 IU/mL) and LoD (0.05 IU/mL). And the observed percentage is greater than the acceptance criteria (87%) in the CLSI document.
The linearity verification result is shown in Supporting Information S1: Figure S1. The best fit linear regression equation for the MAGLUMI HBsAg (CLIA) was y = −4.900 + 0.9234x with R2 = 0.9946, demonstrating a significant correlation between the mean measured and expected HBsAg levels from 0.05 to 250 IU/mL (p < 0.0001).
4 DISCUSSION
In this study, we demonstrate that the MAGLUMI HBsAg (CLIA) has superior seroconversion sensitivity than the ARCHITECT HBsAg, slightly reducing the early window period by 7 days. Meanwhile, the MAGLUMI HBsAg (CLIA) has the capacity to detect a wide range of native and recombinant HBsAg mutants, serotypes, 15 most prevalent HBV subgenotypes (A–H) with relatively high diagnostic sensitivity (100%, N = 411) and detection efficacy. Notably, the sensitivity of the MAGLUMI HBsAg (CLIA) did not comprise the initial diagnostic specificity (99.51%, N = 5306) and had no adverse effects on the detection of samples with potentially cross-reacting substances or unrelated medical conditions. As a result, the MAGLUMI HBsAg (CLIA) has superior sensitivity and specificity, which meets the clinical demands for universal screening and accurate diagnosis of HBV infection.
According to the latest phylogenetic analysis in genotyping of recombinant-free HBV strains, HBV could be classified into 10 (A–J) genotypes and further 24 subgenotypes (A1–A3; B1–B5; C1–C6; D1–D6; and F1–F4).21 Among all 10 genotypes, genotype C (26%), genotype D (22%), E (18%), A (17%), and B (14%) form the global top five most prevalent HBV genotype infections.21 Based on geographical statistics, the most prevalent genotype C is mainly reported in Asia and genotype D is mainly distributed in the Mediterranean area, the Middle East, Russia, and India although genotype D has been found to be prevalent in all continents excluding South America.22 The geographical distribution of genotype E is the West Coast of Africa, Madagascar, Argentina, and Colombia in South America, whereas that of genotype A is Africa, Asia, and Europe.22 Genotype B typically distributes in Asia with a few cases found in Canada and Greenland.22, 23 In this study, 411 positive samples containing genotype A (A1–A3), genotype B (B2–B5), genotype C (C1–C2), genotype D (D1–D4, D7), genotype E, genotype F (F1–F2), genotype G and genotype H have been assessed in parallel to the 1st International reference panel for HBV genotypes for HBsAg assays. The results indicate that the MAGLUMI HBsAg (CLIA) has a high diagnostic sensitivity (100%) and optimal detection efficacy in relation to the most common HBV subgenotypes globally. And the analytical performance verification study confirmed that the MAGLUMI HBsAg (CLIA) is precise, reliable, and a good indicator for HBsAg determination with an excellent linearity from 0.050 to 250 IU/mL. For samples with HBsAg concentrations no larger than 300 000 IU/mL, the MAGLUMI HBsAg (CLIA) could clearly detect samples without high dose hook effect based on the analytical performance study conducted by the manufacturer (Supporting Information S1: Figure S2). These data prove that the MAGLUMI HBsAg (CLIA) is crucial to the HBV universal screening and combating the ever-rising demand for efficient diagnostic testing globally.
With the increasing coverage of vaccination and application of antiviral therapy, the entire HBV genome including HBsAg has emerged constant mutation due to the virus responses, affecting viral antigen presentation, diagnosis, and management of HBV infection.24 When the mutation alters “a” determinant, defined as the immunodominant region of HBsAg, the vaccine-escape mutant would emerge and become the dominant virus whereas the level of wild-type HBsAg would become undetectable.25 Consequently, the probability of immune escape and corresponding failure of HBV vaccination and HBsAg detection would obviously increase, emphasizing the significance of the mutant sensitivity of the HBsAg assay.26 The mutant sensitivity of the MAGLUMI HBsAg (CLIA) has been evaluated by assessing 411 native mutant samples and four recombinant mutant samples from the Diasorin recHBsAg Mutants Panel with corresponding subtype characterizations (Tables 1–3). The results demonstrate that the diagnostic sensitivity of the MAGLUMI HBsAg (CLIA) for detecting the mutant is 100% and the MAGLUMI HBsAg (CLIA) is capable of detecting most of the current native and recombinant HBsAg mutants, which is the necessary performance characteristics for universal screening, diagnosis and management of HBV infection.
The limitations of this study include that the occult HBV infection samples were not involved in our clinical performance evaluation. The occult HBV infection (OBI) refers to the presence of replication-competent HBV DNA, including episomal HBV covalently closed circular DNA, in the liver or the blood of HBsAg negative individuals.27, 28 The gold standard of OBI diagnosis in HBsAg-negative individuals currently is the detection of HBV genomes in liver DNA extracts by liver biopsy since most patients with OBI have undetectable serum HBV DNA levels whereas HBV DNA persists in their liver hepatocytes.27, 29 The prevalence of OBI is reported to be 0.2% in HBsAg negative blood donors and is estimated to be higher in high-endemicity countries, where nucleic acid testing is not implemented for blood screening owing to financial burden and patients are under parenterally transmitted infections such as HCV or HIV.30-32 Therefore, further studies are required to better characterize the clinical performance of the MAGLUMI HBsAg (CLIA) in combination with the MAGLUMI Anti-HCV (CLIA) Test and the MAGLUMI HIV Ab/Ag Combi (CLIA) for detection of OBI, especially in HCV and HIV patients. Moreover, advances in the development of new-generation ultrasensitive HBsAg immunoassays have been reported with significantly improved analytical sensitivity (tenfold) compared with conventional assays and considered to be helpful for the increased identification of HBV carriers with low HBsAg titers and further, control of liver-related complications, prevention of OBI-associated HBV transmission and reactivation.33-35 Although only a small number of cases were involved in the clinical evaluation of these ultrasensitive HBsAg assays and the general increment of HBsAg detection rates varies from 6% to 25% in comparison to conventional assays,33-35 further studies on technical improvement in the analytical sensitivity would be necessary for the management of HBV infection and prevention of HBV transmission, especially in resource-limited settings.
In 2022, the Seventy-Fifth World Health Assembly made the decision to approve the Global health sector strategies on HIV, viral hepatitis, and sexually transmitted infections for the period 2022–2030 (GHSSs) to guide the health sector in implementing strategically focused responses to reach the aim of ending AIDS, viral hepatitis (especially chronic hepatitis B and C) and sexually transmitted infections by 2030.36 This is mainly due to hepatitis B and C have been reported to affect more than 354 million people globally, and at the same time, the co-infection of HBV and HCV as well as the consequent end-stage chronic liver disease is the major cause of death for patients affected by HIV.37, 38 Even though the global hepatitis strategy of WHO has been endorsed by all WHO Member States, researchers have suggested that the current actions are insufficient to reach the goal because of the low diagnosis coverage.6 Moreover, it is recommended that new global strategies and approaches are essential to accomplish the 2030 hepatitis elimination goal by scaling up HBV screening.6 Because the present HBsAg enzyme-linked immunosorbent assays (ELISA) and techniques have relatively long detection time, complex detection process, limited sensitivity and specificity compared with chemiluminescence immunoassay methods (CLIA), HBsAg CLIA detection tests with reasonable price, short detection time, easy performing process and outstanding clinical performance would be helpful for solving the difficult global issue.39 The MAGLUMI HBsAg (CLIA) offers high assay sensitivity, potent subgenotype detection efficacy, and high specificity for the universal screening, diagnosis, and management of HBV infection with reliable, automated, and high throughput platforms.
AUTHOR CONTRIBUTIONS
Lihong Shen: Conceptualization; formal analysis, investigation, writing–review and editing, supervision. Yun Zhang: Conceptualization; resources; methodology; project administration; writing–review and editing. Min Shi: Conceptualization; resources; methodology; project administration; writing–review and editing. Lijia Shao: Data curation; formal analysis; validation; writing–review and editing; supervision. Shengchun Feng: Data curation; formal analysis; validation; writing–review and editing; supervision. Weiting Li: Methodology; formal analysis; writing–original draft; writing–review and editing. Zhonggang Fang: Conceptualization; resources; methodology; project administration; supervision; writing–review and editing. Jun Yin: Methodology; data curation; formal analysis; visualization; project administration; writing–review and editing. Tinghua Li: Conceptualization; resources; project administration; supervision; funding acquisition; investigation; writing–review and editing.
ACKNOWLEDGMENTS
The authors would like to thank Dr Heike Lukhaup for her coordination of the trial and other staff of Biomex GmbH Heidelberg for preparing samples and performing the evaluation experiments. This work was supported by Jinhua Central Hospital (JY2021-1-04) and Jinhua Science and Technology Bureau (2021-4-009). Reagents and technical support were provided by Snibe Diagnostics, Shenzhen New Industries Biomedical Engineering Co. Ltd.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ETHICS STATEMENT
The clinical performance evaluation study was performed by a third-party organization (laboratories of Biomex GmbH) and the assay performance verification study was conducted by the clinical laboratory at Affiliated Jinhua Hospital, Zhejiang University School of Medicine in accordance with the guidelines of the Declaration of Helsinki and in adherence with the principles of Good Clinical Practice. All samples used in this study were residual samples obtaining a general authorization for ethical approval and signed broad consent.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




