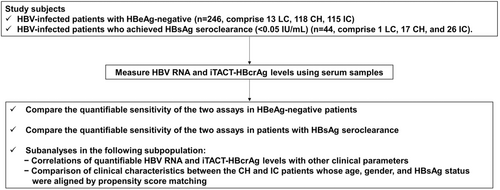
Clinical performance of Circulating HBV RNA and iTACT-HBcrAg Assays in HBeAg-negative and HBsAg-cleared Chronic Hepatitis B Patients
The study protocol was approved by the Institutional Review Board of Nagoya City University (approval number: 60-00-0657).
Abstract
Hepatitis B virus (HBV) RNA and hepatitis B core-related antigen (HBcrAg) have been reported to reflect the transcriptional activity of covalently closed circular HBV DNA. We retrospectively investigated the proportions of quantifiable serum HBV RNA and immunoassay for total antigen including complex via pretreatment-hepatitis B core-related antigen (iTACT-HBcrAg) in chronic hepatitis B patients negative for hepatitis B e antigen (HBeAg) and/or with hepatitis B surface antigen (HBsAg) seroclearance. This study included 246 HBeAg-negative HBV-infected patients, who comprised 13 with liver cirrhosis (LC, the LC group), 118 chronic hepatitis (CH, the CH group), and 115 inactive carriers (IC, the IC group), and 44 patients with HBsAg seroclearance. iTACT-HBcrAg and HBV RNA levels were determined using stored serum samples. Higher proportions of the patients had quantifiable iTACT-HBcrAg than HBV RNA in all groups of HBeAg-negative patients (iTACT-HBcrAg: 84.6%, 90.7%, 35.7%, HBV RNA: 23.1%, 26.3%, 14.8%, for the LC, CH, IC groups). With HBsAg seroclearance (HBsAg <0.05 IU/mL), the proportions of quantifiable samples for HBV RNA were also lower than iTACT-HBcrAg (0% for HBV RNA). Thus, iTACT-HBcrAg was more often detectable than circulating HBV RNA in this study population. Further long-term prospective evaluation of iTACT-HBcrAg is desirable for its utilization in clinical practice.
1 INTRODUCTION
Now, approximately 296 million individuals have persistent hepatitis B virus (HBV) infection worldwide in 2019.1 Several serum HBV biomarkers have been developed, such as HBV DNA, hepatitis B surface antigen (HBsAg), and hepatitis B core-related antigen (HBcrAg).2-5 HBcrAg consists of three proteins encoded by the precore/core region, including hepatitis B e antigen (HBeAg), hepatitis B core antigen (HBcAg), a component of the virion, and a 22 kDa precore protein (p22cr), which is a component of an HBV DNA-negative (empty) particle, and is known to reflect the transcriptional activity of covalently closed circular DNA (cccDNA) within infected hepatocytes.6-8 In clinical practice, HBcrAg has been reported to be useful for early diagnosis of HBV reactivation and prediction of HCC development during administration of nucleot(s)ide analogs (NAs).9, 10 Another HBV marker, serum HBV RNA was also reported to reflect the transcriptional activity of intrahepatic cccDNA.11, 12 Both HBV RNA and HBcrAg levels in sera at the end of treatment were reported to have a better predictive value for off-treatment outcome of NAs.13, 14
In the current practice of HBV, HBsAg seroclearance, which is regarded as a “functional cure,” is a goal of HBV treatment.15 Although HBV-infected patients who achieved HBsAg seroclearance may often have a good prognosis, HBV DNA cannot be completely eradicated because of the persistence of intrahepatic cccDNA and integrated HBV DNA in the hepatocytes, and then the risk of HCC development still remains.15, 16 In fact, we reported that the detection rates of HBcrAg by immunoassay for total antigen including complex via pretreatment (iTACT)-HBcrAg assay varied depending on various phases of chronic HBV infection even after HBsAg seroclearance, and low level of HBcrAg was detected in a case of HCC development after HBsAg seroclearance.17 Thus, it may be important to identify markers reflecting intrahepatic viral activity and their association with the pathogenesis of chronic HBV infection.
Recently, iTACT-HBcrAg (Fujirebio Inc.), which is a fully automated, high-sensitivity chemiluminescent enzyme immunoassay (CLEIA) for HBcrAg detection, has been developed, and the lower limit of quantification (LLOQ) of the assay is 2.1 log U/mL, which has about 10-fold greater sensitivity than the conventional HBcrAg assay (Fujirebio Inc., LLOQ of 3.0 log U/mL).18 On the other hand, an automated, prototype quantitative circulating HBV RNA assay: cobas® HBV RNA using the Roche cobas® 6800/8800 systems (Roche Molecular Diagnostics) has been developed, and the LLOQ of the assay is 10 copies/mL, which also has greater sensitivity than the previous assays.19-21
Therefore, we retrospectively investigated and compared the detection rates of iTACT-HBcrAg and HBV RNA using stored serum samples in HBV-infected patients with HBeAg-negative and/or HBsAg seroclearance, which are considered to have low viral activity and low potential for disease progression.22
2 PATIENTS AND METHODS
2.1 Study design, setting, and study population
The flowchart of this study is shown in Figure 1. This study included 246 HBeAg-negative, HBV-infected patients who presented to the Nagoya City University Hospital, and whose iTACT-HBcrAg levels were measured for clinical examination between December 2022 and September 2023. The 246 patients included 13 with liver cirrhosis (LC, the LC group), 118 with chronic hepatitis (CH, the CH group), and 115 inactive carriers (IC, the IC group) at measurement of iTACT-HBcrAg. None had a history of HCC development. In this study, noncirrhotic and cirrhotic patients who were treated with NAs for more than 1 year were classified as CH and LC respectively, and LC was defined as the presence of the following criteria: nodular liver surface or coarse liver echotexture on ultrasonography (US) and/or clinical features of portal hypertension, including ascites, splenomegaly, esophageal and/or gastric varices, based on imaging tests, such as magnetic resonance imaging (MRI), computerized tomography (CT) and/or US, or thrombocytopenia. IC was defined according to the EASL criteria as follows: being HBeAg negative, hepatitis B e antibody (anti-HBe) positive, HBV DNA < 2000 IU/mL, and ALT < ULN (40 IU/L).15 In addition, we also included 44 HBV-infected patients who presented to the Nagoya City University Hospital between 1994 and 2023 and subsequently achieved HBsAg seroclearance (<0.05 IU/mL), according to conventional HBsAg assays. The 44 patients comprised 1 LC, 17 CH and 26 IC at HBsAg seroclearance. Although we did not pretest and determine the sufficient number of patients for this comparative study, this study include a larger number of patients than the previous study which investigated the detection of HBV RNA in chronic hepatitis B patients.19 Stored serum samples were available from all study patients for the measurement of HBV RNA and iTACT-HBcrAg. Regarding the patients with HBsAg seroclearance, these were the same as those reported previously.17
2.2 Laboratory tests
Serum HBV RNA and iTACT-HBcrAg levels were measured using serum samples that had been stored at −80°C in deep freezer from collection to measurement. The HBV RNA concentrations were quantified by the cobas® HBV RNA, using the Roche cobas® 6800/8800 systems (Roche Diagnostics), with LLOQ 10 copies/mL.23 This assay includes an internal control for nucleic acid recovery, and the amplification target is located the 3' end of HBV transcript, and can detect all viral RNAs expressed from cccDNA.19 Whereas, using stored serum samples or clinical examination, HBcrAg was measured by the iTACT-HBcrAg assay (Fujirebio Inc.) which includes an automated pretreatment process with the pretreatment solution that contained HCl, DEAET (2-(diethylamino) ethanethiol hydrochloride) and detergents, and the immunoassay is completed in approximately 30 min by the fully automated CLEIA system, the LUMIPULSE L2400 (Fujirebio Inc.).18 The detection range of the iTACT-HBcrAg assay is 2.1–7.0 log U/mL. Serum HBV DNA levels were measured by the COBAS AmpliPrep/COBAS TaqMan HBV v2.0 assay (Roche Diagnostics K.K.), according to the manufacturer's instructions. The lower and upper limits of quantitation of the tests were 2.1 log copies/mL and 9.0 log copies/mL until 2017, and 1.3 log IU/mL and 8.2 log IU/mL from 2018, respectively. The measurement method and reagents are the same, but the units have changed in 2018, and the unit change formula is “log IU/mL = log copies/mL −0.76,” according to the manufacturer's instructions. HBsAg was measured by the conventional HISCL HBsAg (Sysmex Corporation) and/or the HBsAg-HQ (Fujirebio Inc.) assays, which both use the CLEIA method. The LLOQ was 0.03 IU/mL for HISCL HBsAg and 0.005 IU/mL for HBsAg-HQ. Serum HBeAg was measured using an automated CLEIA system (HISCL HBeAg; Sysmex Corporation) with a cut-off index of 1.0.
The other hematologic and blood chemistry tests were carried out using standard assays.
2.3 Statistical analysis
Categorical variables were tested by the χ2 test. The data of most parameters were not normally distributed. Noncategorical variables were analyzed using the Mann–Whitney U test or the Kruskal–Wallis test. Correlation coefficients were calculated using the Spearman's correlation coefficient. We performed propensity score matching to align age, gender, and HBsAg levels at baseline between the CH and IC groups. A p value < 0.05 was considered significant in all tests. EZR (Easy R, Saitama Medical Center. Jichi Medical University), which is a modified version of R commander (version 1.61), was used for statistical analysis.24
3 RESULTS
3.1 Baseline clinical and laboratory characteristics of HBeAg-negative patients
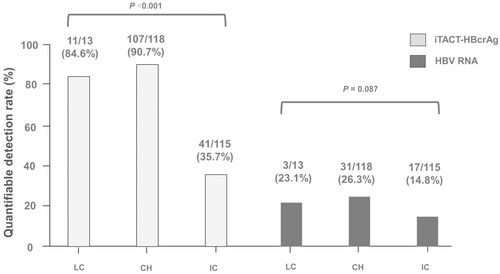
Demographic data of the 246 HBeAg-negative patients are shown in Table 1(A). The LC group was older and had lower platelet counts than the other groups. As for HBsAg levels, the CH group had the highest HBsAg levels of all the groups. The median HBV DNA level was determined in the IC group only, because the patients in the CH and LC groups were treated with NAs. The proportions of quantifiable iTACT-HBcrAg were 84.6% (11/13), 90.7% (107/118), 35.7% (41/115) for the LC, CH, IC groups, respectively. In contrast, the proportions of quantifiable HBV RNA were 23.1% (3/13), 26.3% (31/118), 14.8% (17/115) for the LC, CH, IC groups, respectively. Thus, the proportion of quantifiable iTACT-HBcrAg was lowest in the IC group. In addition, comparing the proportions of quantifiable HBV RNA and iTACT-HBcrAg, iTACT-HBcrAg was more often quantifiable than was HBV RNA, in all the groups (Table 1(A), Figure 2). Table 2 represents the proportions of the four combinations of detection of HBcrAg and HBV RNA in the LC, CH, and IC groups, respectively. The proportion of patients with no detectable HBV RNA but detectable HBcrAg was 61.5% (8/13), 65.3% (77/118), and 24.3% (28/115) in the LC, CH, and IC groups, respectively.
| (A) HBeAg-negative patients | ||||
|---|---|---|---|---|
| HBeAg-negative patients | LC (n = 13) | CH (n = 118) | IC (n = 115) | p Value |
| Age, years | 61 (50‒66) | 54 (46‒62) | 59 (51‒80) | 0.005 |
| Gender, Male/Female | 11/2 | 70/48 | 42/73 | <0.001 |
| Period from the start of the administration of NA, months | 186 (124‒206) | 138 (71‒187) | N.A. | 0.158 |
| Platelet count, ×104/μL | 13.9 (11.9‒17.2) | 21.2 (17.3‒25.5) | 22.1 (18.4‒25.5) | <0.001 |
| ALT, U/L | 18 (14‒21) | 18 (13‒23) | 17 (13‒21) | 0.613 |
| HBV DNA, log IU/mL | N.D. (N.D.‒N.D.) | N.D. (N.D.‒N.D.) | 2.4 (1.6‒3.1) | <0.001 |
| HBsAg, IU/mL | 186.04 (35.04‒275.95) | 472.26 (168.79‒1229.73) | 299.24 (20.43‒1180.02) | 0.009 |
| iTACT-HBcrAg, log U/L | 3.1 (2.4‒3.8) | 3.2 (2.5‒3.7) | 2.1 (2.1‒2.3) | <0.001 |
| The ratio of quantitative detectability of serum HBV RNA, % (number) | 23.1 (3) | 26.3 (31) | 14.8 (17) | 0.087 |
| The ratio of quantitative detectability of iTACT-HBcrAg, % (number) | 84.6 (11) | 90.7 (107) | 35.7 (41) | <0.001 |
| (B) HBV-infected patients at HBsAg seroclearance | ||||
|---|---|---|---|---|
| HBV-infected patients with HBsAg seroclearance | LC (n = 1) | CH (n = 17) | IC (n = 26) | p Value |
| Age at HBsAg loss, years | 65 | 53 (41‒59) | 62 (57‒70) | 0.001 |
| Gender, male/female | 1/0 | 14/3 | 14/12 | 0.128 |
| Platelet count, ×104/μL | 11.1 | 17.9 (14.5‒20.9) | 19.8 (14.2‒22.2) | 0.236 |
| ALT, U/L | 23 | 15 (11‒17) | 15 (10‒32) | 0.465 |
| iTACT-HBcrAg, log U/L | 2.1 | 3.2 (2.3‒3.4) | 2.1 (2.1‒2.2) | <0.001 |
| The ratio of quantitative sensitivity of HBV-DNA, % (number) | 0.0 (0) | 5.9 (1) | 21.7 (5) | 0.462 |
| The ratio of quantitative detectability of serum HBV RNA, % (number) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 1.000 |
| The ratio of quantitative detectability of iTACT-HBcrAg, % (number) | 0.0 (0) | 82.4 (14) | 34.8 (8) | <0.001 |
- Note: Data from all patients are expressed as numbers for categorical data and medians (first–third quartiles) for noncategorical data. Categorical variables were compared between groups using the χ2 test, and noncategorical variables were compared using the Kruskal–Wallis test.
- Abbreviations: ALT, alanine transaminase; CH, chronic hepatitis; HBcrAg, hepatitis B core related antigen; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; IC, inactive carrier; LC, liver cirrhosis; NA, nucleot(s)ide analog; N.A., not applicable; N.D., not detectable.
| HBcrAg/HBV RNA | LC (n = 13) | CH (n = 118) | IC (n = 115) |
|---|---|---|---|
| -/- | 2 (15.4%) | 10 (8.5%) | 70 (60.9%) |
| -/+ | 0 (0%) | 1 (1.0%) | 4 (3.5%) |
| +/- | 8 (61.5%) | 77 (65.3%) | 28 (24.3%) |
| +/+ | 3 (23.1%) | 30 (25.4%) | 13 (11.3%) |
- Note: “-” represents undetectable and “+” represents detectable for HBcrAg and HBV RNA.
- Abbreviations: CH, chronic hepatitis; HBcrAg, hepatitis B core related antigen; HBV, hepatitis B virus; IC, inactive carrier; LC, liver cirrhosis; NA, nucleot(s)ide analog.
3.2 Clinical characteristics of patients with HBsAg seroclearance
The demographic data of the 44 patients at HBsAg seroclearance (<0.05 IU/mL), which include 1 LC, 17 CH, and 26 IC patients are shown in Table 1(B). Serum HBV RNA levels were not quantified in any patients. In contrast, the proportions of quantifiable iTACT-HBcrAg were 0.0% (0/1), 82.4% (14/17), 34.8% (8/26) for the LC, CH, IC groups, respectively. Thus, in HBV-infected patients at HBsAg seroclearance, iTACT-HBcrAg also was more often quantifiable than was HBV RNA.
3.3 Correlations of quantifiable serum HBV RNA and iTACT-HBcrAg levels with other clinical parameters
We next evaluated the correlations of quantifiable serum HBV RNA and iTACT-HBcrAg levels with other clinical parameters. According to a previous report, iTACT-HBcrAg below detection limit was assigned an arbitrary value of 2.0 U/mL, whereas HBV RNA levels detectable but not quantifiable or completely undetectable were assigned arbitrary values of 5 or 1 copies/mL, respectively, for illustrative purposes.23 In the same way, HBV DNA levels detectable but not quantifiable or completely undetectable were assigned arbitrary values of 1.3 or 0 log IU/mL, respectively, for illustrative purposes.
Serum HBV RNA levels were quantified in 31 HBeAg-negative CH patients. As shown in Table 3(A), no factors were significantly correlated with the HBV RNA levels. In contrast, the serum HBV RNA levels were quantified in 17 HBeAg-negative IC patients, and were significantly correlated with the HBV DNA (ρ = 0.570, p = 0.017) and iTACT-HBcrAg levels (ρ = 0.754, p < 0.001).
| (A) Correlations of quantifiable serum HBV RNA levels with other clinical parameters. | ||||
|---|---|---|---|---|
| CH (n = 31) | IC (n = 17) | |||
| Factor | ρ | p Value | ρ | p Value |
| HBV DNA | −0.027 | 0.884 | 0.570 | 0.017 |
| iTACT-HBcrAg | 0.195 | 0.292 | 0.754 | <0.001 |
| HBsAg | −0.271 | 0.141 | 0.444 | 0.075 |
| ALT | −0.178 | 0.336 | 0.451 | 0.069 |
| Platelets | 0.042 | 0.822 | −0.476 | 0.053 |
| (B) Correlations of quantifiable serum iTACT-HBcrAg levels with other clinical parameters. | ||||
|---|---|---|---|---|
| CH (n = 107) | IC (n = 41) | |||
| Factor | ρ | p Value | ρ | p Value |
| HBV DNA | 0.109 | 0.263 | 0.311 | 0.048 |
| HBV RNA | 0.262 | 0.006 | 0.357 | 0.022 |
| HBsAg | 0.202 | 0.037 | 0.323 | 0.039 |
| ALT | 0.090 | 0.358 | −0.021 | 0.899 |
| Platelets | 0.206 | 0.033 | −0.234 | 0.142 |
- Note: Correlation coefficients were calculated using the Spearman's correlation coefficient.
- Abbreviations: ALT, alanine transaminase; CH, chronic hepatitis; HBcrAg, hepatitis B core related antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; IC, inactive carrier.
On the other hand, serum iTACT-HBcrAg levels were quantified in 107 HBeAg-negative CH patients. As shown in Table 3(B), the serum iTACT-HBcrAg levels were significantly correlated with the HBV RNA levels (ρ = 0.262, p = 0.006), HBsAg (ρ = 0.202, p = 0.037), and platelet counts (ρ = 0.206, p = 0.033). In contrast, the serum iTACT-HBcrAg levels were quantified in 41 HBeAg-negative IC patients, and were significantly correlated with HBV DNA (ρ = 0.311, p = 0.048), HBV RNA (ρ = 0.357, p = 0.022), and HBsAg (ρ = 0.323, p = 0.039) levels.
3.4 Comparison of clinical characteristics between the CH and IC patients whose age, gender, and HBsAg status were aligned by propensity score matching
Except for HBV DNA, HBV RNA, and iTACT-HBcrAg, comparing the CH and IC groups, the background parameters age, gender, and HBsAg levels differed significantly between them (Table 1). Subsequently, we carried out propensity score matching and aligned the age, gender, and HBsAg levels in the CH and IC groups and selected 85 patients from each group. However, their HBsAg levels were not fully matched (Table 4). The iTACT-HBcrAg levels were higher in the CH group than the IC group (the median 3.2 vs. 2.1 log IU/L, p < 0.001). The proportion of quantifiable iTACT-HBcrAg was higher in the CH group than the IC group (92.9 [79/85] vs. 32.9 [28/85] %, p < 0.001). In addition, the proportion of quantifiable HBV RNA was also higher in the CH group than the IC group (30.6 [26/85] vs. 14.1 [12/85] %, p = 0.016).
| CH (n = 85) | IC (n = 85) | p Value | |
|---|---|---|---|
| Age, years | 58 (50‒66) | 58 (50‒68) | 0.934 |
| Gender, male/female | 38/47 | 38/47 | 1.000 |
| Period from the start of the administration of NA, months | 152 (96‒191) | N.T. | N.T. |
| Platelet count, ×104/μL | 21.1 (16.8‒25.7) | 22.2 (18.8‒25.5) | 0.090 |
| ALT, U/L | 17 (13‒22) | 17 (13‒21) | 0.901 |
| HBV DNA, log IU/mL | 0.0 (0.0‒0.0) | 2.4 (1.6‒3.1) | <0.001 |
| HBsAg, IU/mL | 485.36 (167.61‒1628.41) | 308.36 (15.34‒941.98) | 0.021 |
| iTACT-HBcrAg, log U/L | 3.2 (2.5‒3.7) | 2.1 (2.1‒2.3) | <0.001 |
| The ratio of quantitative detectability of serum HBV RNA, % (number) | 30.6 (26) | 14.1 (12) | 0.016 |
| The ratio of quantitative detectability of iTACT-HBcrAg, % (number) | 92.9 (79) | 32.9 (28) | <0.001 |
- Note: Data from all patients are expressed as numbers for categorical data and medians (first–third quartiles) for noncategorical data. Categorical and noncategorical variables were compared using the χ2 test and the Mann–Whitney U test, respectively.
- Abbreviations: ALT, alanine transaminase; CH, chronic hepatitis; HBcrAg, hepatitis B core related antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; IC, inactive carrier.
4 DISCUSSION
This study revealed that iTACT-HBcrAg was more detectable than HBV RNA in HBeAg-negative HBV-infected patients. Notably, HBV RNA was not detected in any of the HBV-infected patients at HBsAg seroclearance. Thus, iTACT-HBcrAg may be useful for monitoring the pathogenesis and prognosis of persistent HBV infection in these patients.
Serum HBV RNA levels were significantly correlated with HBV DNA and iTACT-HBcrAg levels in the IC patients who were not treated with antiviral therapy (Table 3), and these results are consistent with a previous report.23 However, no such significant correlations were obtained for the CH patients who were treated with NAs (Table 3). We assume that these markers, HBV RNA, HBV DNA, and iTACT-HBcrAg, were suppressed by antiviral therapy and, as a result, they were no longer correlated.
A previous report showed that deep sequencing of circulating HBV RNA was useful to assess the potential for the selection of drug resistance mutations and subsequent breakthroughs under treatment with NAs.25 However, our data showed that serum HBV RNA was not sufficiently detectable in HBeAg-negative HBV infected patients (in only around 20%), a finding similar to the previous report.23 Therefore, further improvement in the sensitivity of detection serum of HBV RNA is desirable.
In this study, the proportion of quantifiable iTACT-HBcrAg was higher in the LC/CH groups than the IC groups (Table 1). We previously reported that iTACT-HBcrAg could be detected in 54.5% (61/112) of HBeAg-negative IC patients and in 97.5% (157/161) of patients treated with NA at their last visit.18 In addition, it was also reported that iTACT-HBcrAg was detectable in 39.4% (13/33) of IC patients and in 81.5% (23/27) of cirrhosis and chronic hepatitis patients when their HBsAg reached seroclearance (HBsAg <0.05 IU/mL).17 In the present study, iTACT-HBcrAg was detectable in 35.7% (41/115) of HBeAg-negative IC patients, 84.6% (11/13) of cirrhosis and 90.7% (107/118) of chronic hepatitis patients (Figure 2). Thus, the quantitative detectability of iTACT-HBcrAg in this study was not significantly different from the previous studies which used iTACT-HBcrAg assay, and the slight differences may be due to differences in the patients' backgrounds. Above mentioned, serum HBcrAg levels are considered to reflect the activity of intrahepatic cccDNA.8 Therefore, we speculate that the cccDNA is transcriptionally active in the LC/CH group, suggesting the possibility of contributing to the progression of chronic hepatitis B.17 However, this assumption requires further verification.
The limitations of this study are as follows. First, this was a retrospective study. Second, iTACT-HBcrAg and HBV RNA levels were measured at one time point. Third, since the present study used an already commercialized assay, the cobas® HBV RNA only, we were not able to compare the quantifiable sensitivity with other HBV RNA assays. Future studies using other HBV RNA assays are desirable. Fourth, this study did not include an ongoing clinical evaluation of the chronic hepatitis B patients.
5 CONCLUSION
We found that iTACT-HBcrAg was more detectable than HBV RNA in HBV-infected patients negative for HBeAg and/or with HBsAg seroclearance. Further validation of a large number of cases may be necessary, regarding markers in the blood reflecting cccDNA activity under prolonged administration of NAs. In addition, further clinical studies should be conducted to assess the implications of iTACT-HBcrAg compared with HBV RNA assay.
AUTHOR CONTRIBUTIONS
Conception and design: Takanori Suzuki and Kentaro Matsuura. Drafting of manuscript: Takanori Suzuki and Kentaro Matsuura. Acquisition, analysis and interpretation of data: Takanori Suzuki, Kentaro Matsuura, Takako Inoue, Keiko Sasada, Shintaro Ogawa, Takehisa Watanabe, Hayato Kawamura, Kei Fujiwara, Hiromi Kataoka, and Yasuhito Tanaka. Critical revision of the manuscript: Takanori Suzuki and Kentaro Matsuura. Acquisition, analysis and interpretation of data: Takanori Suzuki, Kentaro Matsuura, Takako Inoue, Keiko Sasada, Shintaro Ogawa, Takehisa Watanabe, Hayato Kawamura, and Kei Fujiwara. Study supervision: Hiromi Kataoka and Yasuhito Tanaka.
ACKNOWLEDGMENTS
This research was supported by the Japan Agency for Medical Research and Development (AMED) (Grant Number JP22fk0310518, 23fk0310518h0002).
CONFLICT OF INTEREST STATEMENT
Yasuhito Tanaka: Research funding from Janssen Pharmaceutical K.K., Gilead Sciences, AbbVie GK., GlaxoSmithKline PLC, Fujirebio Incorporation and Sysmex Corporation and received speaker's fees from AbbVie GK., Gilead Sciences, Chugai Pharmaceutical Co., Ltd., ASKA Pharmaceutical Holdings Co., Ltd., OTSUKA Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co., Ltd., and GlaxoSmithKline PLC. Hiromi Kataoka: Honoraria from Takeda Pharmaceutical CO., and Otsuka Pharmaceutical CO. Fees for promotional materials from Eisai CO., and Otsuka Pharmaceutical CO. The remaining authors declare no conflict of interest.
ETHICS STATEMENT
The study protocol was approved by the Institutional Review Board of Nagoya City University (approval number: 60-00-0657). The study was conducted in accordance with the tenets of the Declaration of Helsinki.
Open Research
DATA AVAILABILITY STATEMENT
The authors confirm that the data supporting the findings of this study are available within the article.




