Virological characterization of the 2022 outbreak-causing monkeypox virus using human keratinocytes and colon organoids
Yukio Watanabe, Izumi Kimura, and Rina Hashimoto are cofirst authors.
Abstract
The outbreak-causing monkeypox virus of 2022 (2022 MPXV) is classified as a clade IIb strain and phylogenetically distinct from prior endemic MPXV strains (clades I or IIa), suggesting that its virological properties may also differ. Here, we used human keratinocytes and induced pluripotent stem cell-derived colon organoids to examine the efficiency of viral growth in these cells and the MPXV infection-mediated host responses. MPXV replication was much more productive in keratinocytes than in colon organoids. We observed that MPXV infections, regardless of strain, caused cellular dysfunction and mitochondrial damage in keratinocytes. Notably, a significant increase in the expression of hypoxia-related genes was observed specifically in 2022 MPXV-infected keratinocytes. Our comparison of virological features between 2022 MPXV and prior endemic MPXV strains revealed signaling pathways potentially involved with the cellular damages caused by MPXV infections and highlights host vulnerabilities that could be utilized as protective therapeutic strategies against human mpox in the future.
1 INTRODUCTION
The first documented case of monkeypox virus (MPXV) infection was in the Republic of the Congo in 1970.1 Human mpox cases have since been confirmed in central and western Africa repeatedly. In 2022, however, a new MPXV variant (2022 MPXV) was identified not only in Africa but also in multiple non-African regions such as the United Kingdom, European Union, and the United States.1 As of January 2023, over 84 000 patients have been infected with 2022 MPXV worldwide.
MPXV is a double-stranded DNA virus that belongs to the genus of Orthopoxvirus in the Poxviridae family.2 MPXV strains have been grouped into several clades: Congo Basin clade (clade I), West Africa clade (clade IIa), and 2022 MPXV (clade IIb). A previous analysis of viral genome sequences suggested that MPXV clades IIa and IIb share common virological features.3 It is of critical importance to distinguish between these MPXV strains in case of further outbreaks, however the characterization of 2022 MPXV remains incomplete.
Rashes all over the body, as well as fever, fatigue, severe headache, lymphadenopathy, and muscle aches are often observed in patients infected with MPXV.4, 5 Recovery from human mpox typically occurs after 2−4 weeks, but many severe cases have also been documented and the fatality rate until 2019 was recently reported to be 1%−10%.4 It is also known that MPXV clade I is more lethal and adept at human-to-human transmission than MPXV clade IIa.6, 7 Conversely, 2022 MPXV is currently reported to be relatively less lethal (0.46%) than the endemic MPXV strains. An analysis of the lesion site suggests that 2022 MPXV has a preference for rectal, colon, and genital tissues rather than skin for viral entry and replication.8, 9 However, to our knowledge, no human skin and colon models have been used to date to characterize MPXV infections.
Various cells and animal models have been used to characterize MPXV. For instance, BSC-40, LLC-MK2, and Vero cells10, 11 have been used for MPXV infection experiments and drug development research in vitro. However, these cell models are monkey-derived cells and are not suitable for analyzing host responses to MPXV in humans. To understand the virological features of MPXV in depth, it is essential to use in vitro cell models (e.g., organoids) that faithfully reproduce human pathophysiology.
In this study, we used three MPXV strains, clade I, IIa, and 2022 MPXV, to characterize the virological features of 2022 MPXV. Using human keratinocytes and induced pluripotent stem (iPS) cell-derived colon organoids to model human skin and colon—the presumed primary sites of MPXV infection—we examined and contrasted organ tropism, host immune response, and tissue damage caused by infections with MPXV clades I, IIa, and 2022 MPXV.
2 MATERIALS AND METHODS
2.1 Resource availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
2.2 NHEK
NHEK-Ad (Normal Human Epidermal Kerationocytoes-Adult; Cat# 00192627; Lonza) were cultured with KGM-Gold Keratinocyte Growth Medium BulletKit (Cat# 00192060; Lonza). NHEK-Ad were seeded on 24-well cell culture plates (2.0 × 104 cells/2 cm2) and cultured for 7 days.
2.3 Human iPS cell-derived colon organoids
The iPS cell line, 1383D6, was maintained on 0.5 μg/cm2 recombinant human laminin 511 E8 fragments (iMatrix-511; Cat# 892 012; Nippi) with StemFit AK02N medium (Cat# RCAK02N; Ajinomoto) containing 10 μM Y-27632 (Cat# 034-24024; FUJIFILM Wako Pure Chemical). To passage the cells, iPS cell colonies were treated with TrypLE Select Enzyme (Cat# 12563029; Thermo Fisher Scientific) for 10 min at 37°C. After centrifugation, cells were seeded on Matrigel® Growth Factor Reduced Basement Membrane (Cat# 354230; Corning)-coated cell culture plates (2.0 × 105 cells/4 cm2) and cultured for 2 days. To perform definitive endoderm differentiation, human iPS cells were treated with 100 ng/mL Activin A (Cat# 338-AC-01M; R&D Systems) and 10 μM Y-27632 in RPMI1640 medium (Cat# R8758-500; Sigma-Aldrich) supplemented with 1× B-27 Supplement Minus Vitamin A (Cat# 12587001; Thermo Fisher Scientific), 1× GlutaMAX (Cat# 35050-079; Thermo Fisher Scientific), and 1× penicillin−streptomycin for 3 days. To perform hindgut differentiation, the cells were treated with 200 ng/uL FGF2 (Cat# 160-0010-3; Katayama Chemical Industries) in RPMI1640 medium supplemented with 1× B-27 Supplement Minus Vitamin A, 1× GlutaMAX, and 1× penicillin−streptomycin for 4 days. To generate colon organoids, the cells were dissociated and embedded in Matrigel Growth Factor Reduced Basement Membrane to generate organoids. To perform colonic differentiation, the cells were treated with 3 μM CHIR99021 (Cat# 034-23103; FUJIFILM Wako Pure Chemical), 0.5 μM A-83-01 (Cat# 035-24113; FUJIFILM Wako Pure Chemical), 50 ng/mL Noggin (Cat# 120-10C; PeproTech), 30 ng/uL Forskolin (Cat# 063-02193; FUJIFILM Wako Pure Chemical), and 50 ng/mL EGF (Cat# AF-100-15; PeproTech) in advanced DMEM/F12 supplemented with 1× N2 (Cat# 141-08941; FUJIFILM Wako Pure Chemical), 1× B-27 Supplement Minus Vitamin A, 1× GlutaMAX, 0.05% bovine serum albumin, and 1× penicillin−streptomycin for 13 days. To perform the infection experiments, the colon organoids were recovered from Matrigel and the suspension of colon organoids (small free-floating clumps) was seeded onto Matrigel-coated 24-well cell culture plates (1.0 × 105 cells/2 cm2) and cultured for 3 days.
2.4 MPXV preparation
MPXV infection experiments were performed in a biosafety level 3 (BSL3) facility. Three MPXV strains, Zr-599 (Congo Basin strain, clade I), Liberia (West African strain, clade IIa), and the 2022 outbreak strain (MPXV/human/Japan/Tokyo/TKY220091/2022, clade IIb, Genbank accession no. LC722946.1), were propagated using VeroE6 cells (kindly gifted by Tokyo Metropolitan Institute of Public Health). Zr-599 and Liberia strains were kindly gifted by National Institute of Infectious Diseases. The 2022 outbreak strain was kindly gifted by Tokyo Metropolitan Institute of Public Health.
To prepare working MPXV stock, VeroE6 cells (5 million cells) were seeded in a T-75 flask. Next day, the culture medium was refreshed with DMEM (low glucose) (Cat# D6046-500ML; Sigma-Aldrich) containing 2% fetal bovine serum (FBS; Cat# 172012-500ML; Sigma-Aldrich), and 1× penicillin−streptomycin (Cat# P4333-100ML; Sigma-Aldrich), and seed virus was inoculated. At 3 or 4 dpi, the infected flask including culture supernatant and cells were frozen at –80°C. Before use, the flask was placed at room temperature to facilitate thawing. The thawed medium in the flask was harvested and centrifuged, and the supernatant was collected as a working MPXV stock.
2.5 MPXV titration
Virus infectious titers were determined by plaque assay.31 One day before infection, VeroE6 cells (0.2 million cells) were seeded into a 12-well plate and infected with MPXV. Before infection, the culture medium was changed to DMEM (low glucose) containing 2% FBS, and 1% penicillin−streptomycin. The cells were incubated at 37°C in a fully-humidified atmosphere of 5% CO2. At 3 dpi, the culture medium was removed, and the cells were washed with PBS once and fixed with 4% paraformaldehyde phosphate (Cat# 09154-85; Nacalai Tesque). The fixed cells were washed with tap water, dried, and stained with a staining solution (0.1% methylene blue [Cat# 22412-14; Nacalai Tesque] in water) for 30 min. The stained cells were washed with tap water and dried, and the number of plaques was measured to calculate plaque forming unit.
2.6 Quantification of viral DNA copy number of the cell culture supernatant
The cell culture supernatant was mixed with an equal volume of 2× DNA lysis buffer (distilled water containing 2% Triton X-100, 50 mM KCl, 100 mM Tris-HCl [pH 7.4], and 40% glycerol) and incubated at room temperature for 10 min. The mixture was diluted 10 times with distilled water. Viral DNA was quantified using a Probe qPCR Mix (Cat# RR391A; Takara Bio) with 5’-GGA AAA TGT AAA GAC AAC GAA TAC AG-3’ as the forward primer and 5’-GCT ATC ACA TAA TCT GGA AGC GTA-3’ as the reverse primer, and 5’-FAM-AAG CCG TAA TCT ATG TTG TCT ATC GTG TCC-TAMRA-3’ as the probe. To quantify viral DNA copy number, a plasmid encoding MPXV G2R gene was used as the standard.
2.7 Quantification of viral DNA copy number in the cell
Total DNA was isolated using DNeasy Blood & Tissue Kit (Cat# 69506; QIAGEN). For quantification of MPXV DNA, Monkeypox virus qPCR Detection Kit (Cat# RC420A; Takara Bio) was used on a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific). Standard curves were prepared using plasmids encoding MPXV G2R gene.
2.8 Quantitative PCR
Total RNA was isolated using ISOGENE (Cat# 319-90211; NIPPON GENE). cDNA was synthesized using 500 ng of total RNA with the Superscript VILO cDNA Synthesis Kit (Cat# 11754250; Thermo Fisher Scientific). Real-time RT-PCR was performed with SYBR Green PCR Master Mix (Cat# 4385614; Thermo Fisher Scientific) using a StepOnePlus real-time PCR system, QuantStudio 1, or QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific). The relative quantification of target mRNA levels was performed using the method. The values were normalized to the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The primer sequences are summarized in Supporting Information: Table S1.
2.9 Immunofluorescence staining
For immunofluorescence staining of human iPS cell-derived colon organoids, the cells were fixed with 4% paraformaldehyde in PBS at 4°C. Human iPS cell-derived colon organoids were harvested and used for the preparation of paraffin Section (5 μm). The sections were permeabilized using Tris Buffered Saline with 0.1%-Tween 20 Detergent (Cat# 34874-91; Nacalai Tesque) for 10 min and blocked using Blocking One Histo (Cat# 06349-64; Nacalai Tesque) for 45 min. The sections were incubated in Tris Buffered Saline with 0.1%-Tween 20 Detergent with or without primary antibodies overnight at 4°C. The sections were washed with PBS and incubated in Tris Buffered Saline with 0.1%-Tween 20 Detergent containing Alexa Fluor 488- or 594-conjugated secondary antibodies for 45 min. The sections were finally washed and mounted with Fluoro-KEEPER Antifade Reagent, Non-Hardening Type with DAPI (Cat# 12593-64; Nacalai Tesque) and analyzed using the BZ-X700 (Keyence Corporation). The antibodies are summarized in Supporting Information: Table S2.
2.10 RNA sequencing
Total RNA was isolated from cells using ISOGENE. RNA integrity was assessed using a 2100 Bioanalyzer (Agilent Technologies). Library preparation was performed using the TruSeq Stranded mRNA Sample Prep Kit (Cat# 20020594; Illumina) according to the manufacturer's instructions. Sequencing was performed on an Illumina NextSeq. 550. The fastq files were generated using bcl2fastq-2.20. Adapter sequences and low-quality bases were trimmed from the raw reads using Cutadapt ver v3.4.32 The trimmed reads were mapped to human reference genome sequences (hg38) using STAR ver 2.7.9a33 with the GENCODE (release 36, GRCh38.p13)34 gtf file. The raw counts were calculated using htseq-count ver. 0.13.535 with the GENCODE gtf file. Gene expression levels were determined as transcripts per kilobase million (TPM) values with DESeq. 2 v1.30.1.36 Raw data generated from this study were submitted under Gene Expression Omnibus accession number GSE219036.
2.11 Phylogenetics analysis of MPXV
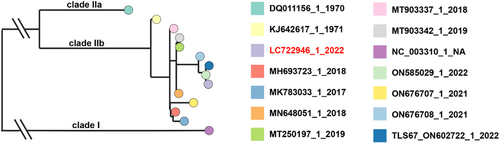
A phylogenetic analysis of MPXV was performed based on core SNP alignment (906 variant positions) retrieved from a rapid alignment (parsnp version 1.2) of the available MPXV genome sequences, with reference genomes, Zaire-96-I-16 (Genbank accession no. NC_003310.1, MPXV clade I) and Liberia_1970_184 (Genbank accession no. DQ011156.1, MPXV clade IIa). Phylogenetic data visualization was performed with Microreact (https://microreact.org/).
2.12 Statistical analyses
Statistical significance was evaluated using unpaired two-tailed Student's t-test, one-way analysis of variance (ANOVA) followed by Dunnett or Tukey post hoc tests, or two-way ANOVA followed by Tukey post hoc tests. Statistical analyses were performed using GraphPad Prism 9. Details are described in the figure legends.
3 RESULTS
3.1 MPXV is more likely to infect keratinocytes than colon organoids
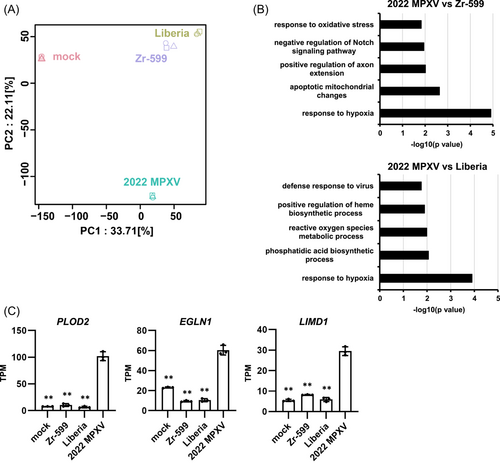
Three MPXV strains were used in this study to characterize MPXV infections: Zr-599 MPXV (Congo Basin monkeypox virus, clade I), Liberia MPXV (West African monkeypox virus, clade IIa), and an isolate from the 2022 outbreak (strain TKY220091, clade IIb; referred to as “2022 MPXV” hereafter). 2022 MPXV is classified as a clade IIb strain (lineage B.1) and phylogenetically distinct from prior endemic MPXV strains (Figure 1). In this study, normal human epidermal keratinocytes (NHEK) and colon organoids were infected separately with the above MPXV strains and cultured for 3 days (Figure 2A). MPXV genomes were detected in the cell culture supernatants of infected keratinocytes and colon organoids at 1, 2, and 3 day(s) postinfection (dpi) (Figure 2B). In the cell culture supernatant of 2022 MPXV-infected keratinocytes, the virus copy number increased 16.7-fold from 1 dpi to 3 dpi (Figure 2B, left), while the increase observed during the same period was only 1.4-fold for infected colon organoids (Figure 2B, right). At 3 dpi, viral copy numbers in the cell culture supernatant of infected keratinocytes were 41.6, 7.0, and 8.2 times higher for Zr-599 MPXV, Liberia MPXV, and 2022 MPXV than those of infected colon organoids, respectively. We did not observe any considerable variation between the MPXV clades in the quantity of viral genome in the culture supernatant of infected colon organoids. On the other hand, the viral copy number in the culture supernatant of Zr-599 MPXV-infected keratinocytes was higher than that of Liberia MPXV-infected keratinocytes. In addition, MPXV DNA and mRNA were detected in infected keratinocytes and colon organoids (Figure 2C,D). The intracellular MPXV mRNA expression in infected keratinocytes was higher than that in infected colon organoids (Figure 2D). Consistent with this, a Circos plot showing the distribution of reads along the MPXV genome indicated that keratinocytes were more efficiently infected than colon organoids (Figure 2E). Interestingly, the amounts of Zr-599 MPXV DNA (Figure 2C) and mRNA (Figure 2D) were slightly higher than those detected from Liberia MPXV- and 2022 MPXV-infected colon organoids (Zr-599 MPXV/Liberia MPXV = 1.3 and 1.6, Zr-599 MPXV/2022 MPXV = 3.1 and 4.8) suggesting that Zr-599 MPXV had a higher infection efficiency in colon organoids than Liberia MPXV and 2022 MPXV. These results suggest that 2022 MPXV shares similar infection tropism with prior endemic MPXV strains.
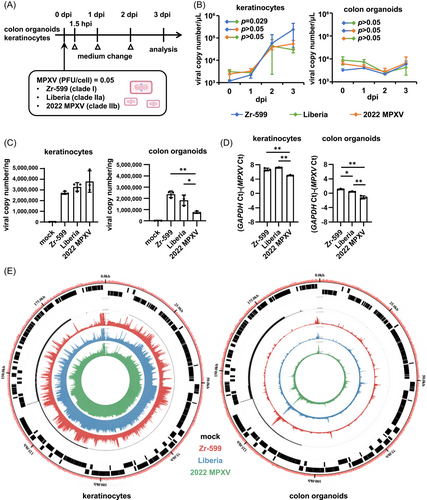
In addition, infection experiments with the three MPXV clades were performed using VeroE6 cells, which are widely used for MPXV research. Although there was almost no viral genome present in the cell culture supernatant of infected VeroE6 cells, both intracellular MPXV DNA and mRNA were detected (Supporting Information: Figures S1A, S1B). MPXV mRNA expression detected in infected VeroE6 cells was similar to infected colon organoids, suggesting that the infection efficiency was similar between VeroE6 cells and colon organoids, but much lower than in keratinocytes.
3.2 MPXV infection causes cellular damage in keratinocytes
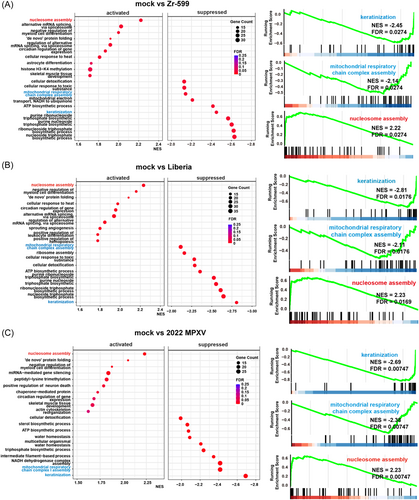
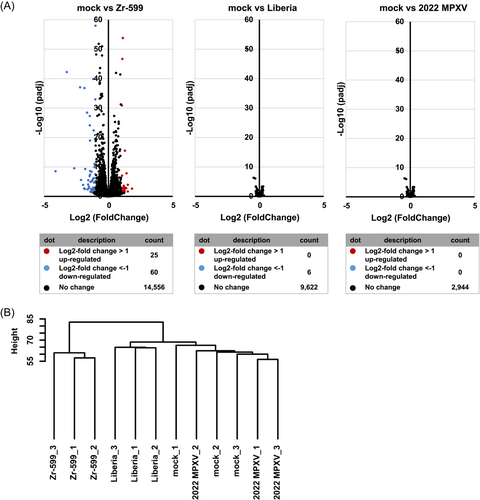
As shown in Figure 3A, cytopathic effects of Zr-599 MPXV, Liberia MPXV, or 2022 MPXV infections were observed in keratinocytes. In addition, most keratinocytes became KRT10 (a keratinocyte marker)-negative following Zr-599 MPXV, Liberia MPXV, and 2022 MPXV infections (Figure 3B). We next performed RNA-seq analysis to examine the host response by keratinocytes caused by MPXV infection. Volcano plots in Figure 3C show that infections with Zr-599 MPXV, Liberia MPXV, or 2022 MPXV significantly altered the expression levels of 938, 1529, and 968 genes in keratinocytes, respectively. Cluster analysis suggested that the gene expression profile of keratinocytes infected with 2022 MPXV was different from those of keratinocytes infected with Zr-599 MPXV or Liberia MPXV (Figure 3D). In Figure 4, dot plots of the top 10 significantly enhanced or decreased gene sets from Gene Set Enrichment Analysis (GESA) of Zr-599 MPXV- (Figure 4A), Liberia MPXV- (Figure 4B), or 2022 MPXV- (Figure 4C) infected keratinocytes compared with uninfected keratinocytes (mock) are shown. The expression levels of genes in the “keratinization” and “mitochondrial respiratory chain complex assembly” categories were decreased in Zr-599 MPXV-, Liberia MPXV-, and 2022 MPXV-infected keratinocytes (Figure 4). Consistently, previous histologic and immunohistochemical analyzes in human mpox patients have also demonstrated keratinocyte dysfunction, including necrosis of keratinocytes.15 These results suggest that efficient proliferation of MPXV in keratinocytes disrupts their normal functions. Moreover, genes in the “nucleosome assembly” category were increased in Zr-599 MPXV-, Liberia MPXV-, and 2022 MPXV-infected keratinocytes (Figure 4). A previous study, which performed microarray analysis in MPXV-infected MK2, showed that the majority of modulated histone-encoding genes were significantly elevated in expression.16 Taken together, these results suggest that MPXV infections may alter the regulation of chromatin structure in host cells.

Interestingly, both cluster analysis (Figure 3D) and principal component analysis (PCA) (Figure 5A) revealed that infection with 2022 MPXV led to gene expression profile changes that were unique to this strain. To further explore this finding, we extracted the set of genes whose expression levels were specifically altered by 2022 MPXV infection for an in-depth examination. As shown in Figure 5B, we performed DAVID-based gene ontology (GO) enrichment analysis on this gene set. The top five significantly enriched GO terms in 2022 MPXV-infected keratinocytes compared with Zr-599 MPXV- and Liberia MPXV-infected keratinocytes are listed in Figure 5B. Interestingly, the top significantly enriched GO term was “response to hypoxia.” We confirmed by RT-PCR that the expression levels of Egl-9 Family Hypoxia Inducible Factor 1 (EGLN1)17 and LIM Domain Containing 1 (LIMD1),18 which are closely related to hypoxia, were increased only by 2022 MPXV infection (Figure 5C). In addition, a hypoxia-induced gene, procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (PLOD2),19 was also specifically upregulated by 2022 MPXV infection (Figure 5C). These results suggest that skin lesions induced by MPXV may be associated with abnormal hypoxic signaling and collagen production capacity. Altogether, our findings raise the possibility that 2022 MPXV induces skin lesions via mechanisms different from those of prior endemic MPXV strains.
3.3 Zr-599 MPXV induces cellular damage in colon organoids
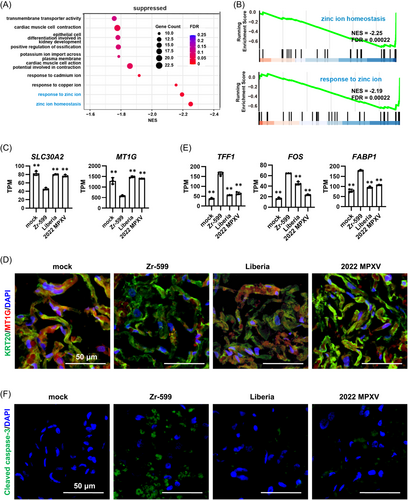
We next turned our attention to the MPXV-infected colon organoids and characterized how they responded to MPXV infections via RNA-seq analysis. Volcano plots in Figure 6A show that Zr-599 MPXV, Liberia MPXV, and 2022 MPXV infections in colon organoids, in contrast to keratinocytes, did not induce much changes in gene expression profile. Nonetheless, we did identify 85 genes whose expression levels were changed more than twofold in Zr-599 MPXV-infected colon organoids. Cluster analysis suggested that the gene expression profile of colon organoids infected with Zr-599 MPXV differs from those of colon organoids infected with Liberia MPXV or 2022 MPXV (Figure 6B). As such, we performed GESA on the set of genes whose expression was decreased in colon organoids by Zr-599 MPXV infection (Figure 7A). The top two significantly enriched GO terms were “response to zinc ion” and “zinc ion homeostasis.” Barcode plots similarly showed that the gene expression levels of the “response to zinc ion” and “zinc ion homeostasis” categories were decreased in Zr-599 MPXV-infected colon organoids (Figure 7B). It is known that zinc absorption and excretion capacities are deeply involved in intestinal homeostasis.20 Both solute carrier family 30 member 2 (SLC30A2) and metallothionein 1G (MT1G) are essential for maintaining an appropriate concentration of zinc, but their expressions were significantly decreased in Zr-599 MPXV-infected colon organoids (Figure 7C). Moreover, immunostaining analysis showed that the protein level of MT1G was consistently decreased in Zr-599 MPXV-infected colon organoids (Figure 7D). Previous studies have reported SLC30A2 gene depletion21, 22 or reduced MT1G expression23, 24 to induce intestinal disorders. Importantly, Zr-599 MPXV infection induced gene expression of intestinal inflammation markers, such as trefoil factor 1 (TFF1), fos proto-oncogene, AP-1 transcription factor subunit (FOS), and fatty acid binding protein 1 (FABP1), in colon organoids (Figure 7E). Immunostaining analysis revealed that cleaved Caspase-3 expression was highest in Zr-599 MPXV-infected colon organoids, suggesting that Zr-599 MPXV infection induced apoptosis in colon organoids (Figure 7F). Consistent with this, Zr-599 MPXV, but not Liberia MPXV, has been reported to induce severe intestinal damage in nonhuman primates.7 These results suggest that Zr-599 MPXV may induce intestinal disorders by disturbing zinc homeostasis in the colon.
4 DISCUSSION
In this study, we used human keratinocytes and colon organoids to clarify the difference between 2022 MPXV and two endemic MPXV strains in terms of infection efficiency and host responses. Although there was little difference in MPXV replication efficiency among the three MPXV strains, MPXV replication was much more productive in keratinocytes than in colon organoids. RNA-seq analyses further revealed changes in gene expression profile unique to 2022 MPXV-infected keratinocytes and Zr-599 MPXV-infected colon organoids.
Notably, we found that 2022 MPXV infection strongly induced hypoxia signaling in keratinocytes (Figure 5) and Zr-599 MPXV infection decreased the expression of genes essential for maintaining zinc homeostasis in colon organoids (Figure 7). It remains to be determined whether induction of hypoxia signaling and abnormal zinc homeostasis contribute to keratinocyte and colonic dysfunction, respectively. Furthermore, it is necessary to confirm whether these phenomena are also observed in keratinocytes and the colon of human mpox patients. Although to our knowledge, no drugs have been developed to specifically target host proteins, we hope that our findings will lead to the development of drugs that can protect host cells against such dysfunctions. By conducting mechanistic analysis of human mpox using our MPXV infection models comprised of human keratinocytes or iPS cell-derived colon organoids, the virological characteristics of each MPXV clade may be clarified in greater detail in future studies.
We found the MPXV infection efficiency in keratinocytes to be more than ten times greater than that in colon organoids (Figure 2B,E). Consistent with this, in human mpox patients, no other organ is known to possess a higher viral load than the skin, thus highlighting it as the main site of MPXV infection.25 Differences in the expression levels of viral receptors and related proteins required for MPXV infection could potentially explain this observed organ tropism. However, to date, the full suite of receptors and accessory proteins used by MPXV to infect host cells has yet to be identified. In the future, by deleting candidate genes for receptors related to MPXV infection in human keratinocytes or colon organoids, it will be evident whether they truly contribute to MPXV infection. In addition to the skin, MPXV was detected in throat swabs and semen of human mpox patients26-28 and a recent report showed that MPXV can infect human iPS cell-derived astrocytes and neural progenitor cells29; altogether suggesting that MPXV likely has the potential to infect and replicate in these and other organs as well.
In this study, we succeeded in evaluating both viral infection and host responses using human keratinocytes and iPS cell-derived colon organoids. Using these models, it may be possible to develop new therapeutic agents against human mpox. Although therapeutic drugs, which target MXPV proteins, such as tecovirimat, cidofovir, and brincidofovir have already been developed,30 their antiviral effects in human cells have not been fully examined. In addition, MPXV replication efficiency was higher in human keratinocytes than in Vero cells, thus allowing for a more sensitive evaluation of antiviral drugs. Furthermore, it may be feasible to evaluate not only antiviral drugs but also drug candidates that can ameliorate tissue damage in the skin and colon using human keratinocytes and iPS cell-derived colon organoids, respectively. Like severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), MPXV may acquire novel mutations constantly, thus necessitating the development of diverse drugs. Both colon organoids and keratinocytes can be cultured in a 96-well plate format, making them suitable for large-scale high-throughput drug screens. We hope that our findings will accelerate the understanding of human mpox infections and facilitate drug discovery research for its treatment.
G2P-JAPAN CONSORTIUM
Jumpei Ito, Rigel Suzuki, Keiya Uriu, Yukari Itakura, Jiri Zahradnik, Sayaka Deguchi, Lei Wang, Spyros Lytras, Tomokazu Tamura, Izumi Kida, Hesham Nasser, Maya Shofa, MST Monira Begum, Masumi Tsuda, Yoshitaka Oda, Shigeru Fujita, Kumiko Yoshimatsu, Hayato Ito, Naganori Nao, Hiroyuki Asakura, Mami Nagashima, Kenji Sadamasu, Kazuhisa Yoshimura, Yuki Yamamoto, Tetsuharu Nagamoto, Gideon Schreiber, Akatsuki Saito, Keita Matsuno, Shinya Tanaka, Takasuke Fukuhara, Terumasa Ikeda, Saori Suzuki, Marie Kato, Zannatul Ferdous, Hiromi Mouri, Kenji Shishido, Naoko Misawa, Izumi Kimura, Yusuke Kosugi, Pan Lin, Mai Suganami, Mika Chiba, Ryo Yoshimura, Kyoko Yasuda, Keiko Iida, Naomi Ohsumi, Adam P. Strange, Daniel Sauter, So Nakagawa, Jiaqi Wu, Takao Hashiguchi, Tateki Suzuki, Kanako Kimura, Jiei Sasaki, Yukari Nakajima, Hisano Yajima, Kotaro Shirakawa, Akifumi Takaori-Kondo, Kayoko Nagata, Yasuhiro Kazuma, Ryosuke Nomura, Yoshihito Horisawa, Yusuke Tashiro, Yugo Kawa, Takashi Irie, Ryoko Kawabata, Ryo Shimizu, Otowa Takahashi, Kimiko Ichihara, Chihiro Motozono, Mako Toyoda, Takamasa Ueno, Yuki Shibatani, Tomoko Nishiuchi.
AUTHOR CONTRIBUTIONS
Yukio Watanabe: RNA-seq analysis, analysis of infected in vitro models, preparation of the manuscript. Izumi Kimura: Infection experiments, data analyses. Rina Hashimoto: Generation of in vitro models, analysis of infected in vitro models, preparation of the manuscript. Ayaka Sakamoto: Generation of in vitro models, analyzed infected in vitro models. Naoko Yasuhara: Analysis of infected in vitro models. Takuya Yamamoto: RNA-seq analysis. The Genotype to Phenotype Japan (G2P-Japan) Consortium: Contribute to the project administration. Kei Sato: Research design, review and editing of the manuscript, funding acquirement, final approval. Kazuo Takayama: Research design, review and editing of the manuscript, funding acquirement, final approval.
ACKNOWLEDGMENTS
We would like to thank all members belonging to The Genotype to Phenotype Japan (G2P-Japan) Consortium. We thank Dr. Kelvin Hui (Kyoto University) for critical reading of the manuscript, Ms. Kazusa Okita and Ms. Satoko Sakurai (Kyoto University) for technical assistance with the RNA-seq experiments, and Ms. Natsumi Mimura (Kyoto University) for technical assistance. This research was supported by the iPS Cell Research Fund, the Japan Agency for Medical Research and Development (AMED) (JP21gm1610005, JP21fk0108425, JP223fa727002, JP223fa627001).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The RNA-seq data that support the findings of this study are openly available in Gene Expression Omnibus at https://www.ncbi.nlm.nih.gov/geo/, reference number GSE219036. Other data that support the findings of this study are available from the corresponding authors upon request.