A substrate for a cell free in vitro assay system to screen drugs targeting trypsin like protease-based cleavage of SARS-CoV-2 spike glycoprotein and viral entry
Prem Praksh Tripathi is co-corresponding author.
Abstract
Host proteases trypsin and trypsin-like proteases have been reported to facilitate the entry of coronavirus SARS-CoV-2 in its host cells. These protease enzymes cleave the viral surface glycoprotein, spike, leading to successful cell surface receptor attachment, fusion and entry of the virus in its host cell. The spike protein has protease cleavage sites between the two domains S1 and S2. Since the cleavage site is recognized by the host proteases, it can be a potential antiviral therapeutic target. Trypsin-like proteases play an important role in virus infectivity and the property of spike protein cleavage by trypsin and trypsin-like proteases can be used to design assays for screening of antiviral candidates against spike protein cleavage. Here, we have documented the development of a proof-of-concept assay system for screening drugs against trypsin/trypsin-like proteases that cleave spike protein between its S1 and S2 domains. The assay system developed uses a fusion substrate protein containing a NanoLuc luciferase reporter protein, the protease cleavage site between S1 and S2 domains of SARS-CoV-2 spike protein and a cellulose binding domain. The substrate protein can be immobilized on cellulose via the cellulose binding domain of the substrate. When trypsin and trypsin-like proteases cleave the substrate, the cellulose binding domain remain bound to the cellulose and the reporter protein is dislodged. Reporter assay using the released reporter protein is the read out of the protease activity. We have demonstrated the proof-of-concept using multiple proteases like trypsin, TMPRSS2, furin, cathepsin B, human airway trypsin and cathepsin L. A significant increment in fold change was observed with increasing enzyme concentration and incubation time. Introduction of increasing amounts of enzyme inhibitors in the reaction reduced the luminescent signal, thus validating the assay. Furthermore, we used SDS-PAGE and immunoblot analyses to study the cleavage band pattern and re-confirm the cleavage for enzymes tested in the assay. Taken together, we have tested an in-vitro assay system using the proposed substrate for screening drugs against trypsin like protease-based cleavage of SARS-CoV-2 spike glycoprotein. The assay system can also be potentially used for antiviral drug screening against any other enzyme that might cleave the used cleavage site.
1 INTRODUCTION
The surface glycoprotein, spike, of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the main surface antigen of the virus which it interacts with host receptors mediating virus entry. The SARS-CoV-2 spike protein has two subunits: S1 and S2. The S1 subunit is involved in host receptor recognition and attachment via the receptor binding domain (RBD) while the S2 subunit is involved in viral entry by helping in fusion of the viral and host cell membranes after the spike is cleaved by host cell proteases.1-4
Angiotensin-converting enzyme 2 (ACE2) serves as the main receptor which enables the virus to enter the lung epithelial cells and causes the disease.1, 3, 5 Once the virus attaches to the receptor, the spike protein undergoes cleavage at its two potential sites (S1/S2 and S2') by the host cell proteases. This brings about a conformational change enabling the fusion process further.4, 6 As compared to the other coronaviruses, SARS-CoV-2 spike is more susceptible to cleavage by the host proteases due to the presence of a multibasic arginine rich sequence at the S1/S2 site.1, 3 Studies have shown multiple host proteases to be involved in facilitating the virus entry notably human airway trypsin like protease (HAT), Type II transmembrane serine protease (TMPRSS2), furin and cathepsins.1-3, 5, 7-10 Furin has been shown to cleave SARS-CoV-2 spike protein at the S1/S2 site but the role of the S2' site in getting recognized by these proteases remains to be elucidated.11
Previous studies on SARS-CoV have manifested the role of HAT in facilitating cleavage of the spike protein which in turn may help in virus entry in host cells.12 Also, recent investigations on SARS-CoV-2 have revealed the significance of trypsin and HAT in virus entry by cell-cell fusion assay where the fusogenic activity of the protein was enhanced in the presence of these proteases.7 In another study, SARS-CoV-2 infection was also shown to be enhanced in the presence of exogenous trypsin indicating the possible role of trypsin or trypsin-like proteases in SARS-CoV-2 infection.9 A serine protease, TMPRSS2 has been shown to play a pivotal role in increasing the infectivity of the virus by processing the spike protein.1, 8 Earlier, TMPRSS2 has been shown to be important in infection of other related human coronaviruses; SARS-CoV and MERS-CoV by cleaving the surface glycoprotein after receptor recognition.13-15 Studies on MERS-CoV have shown S2' cleavage site to be important for the recognition process by TMPRSS2 and enabling virus entry.15 TMPRSS2 is a monobasic site-specific protease that cleaves after a single arginine or lysine residue and in MERS-CoV spike the S2'site harbors arginine at two different positions forming a monobasic site suitable for TMPRSS2 cleavage.15 Furin is another such host protease that has been well studied in context of SARS-CoV-2 spike cleavage at the S1/S2 site, promoting the virus infection.2, 3 Cathepsin B/L are also known to promote SARS-CoV-2 entry in TMPRSS2 deficient cells indicating the role of multiple host proteases in the cleavage process depending on cell types and abundancy.1, 2, 10
The long term COVID-19 pandemic necessitated development of antiviral therapeutics that can help in managing the infection. Repurposing drugs and developing new antivirals that can inhibit the viral entry are important research areas. In this study, we have focused on the development of an in vitro nano luciferase-based assay using a fusion protein (referred to as the substrate in this article) containing three different protein components; NanoLuc luciferase (NL), SARS-CoV-2 spike sequence (CS) (protease cleavage area between S1 and S2 domains) and cellulose binding domain (CBD). The NanoLuc luciferase is a luminescent protein that undergoes a bioluminescent reaction on its substrate (furimazine) in the presence of oxygen. The protein is active both in its fused and free state, but the luminescent signal increases many fold when the enzyme is free. The SARS-CoV-2 spike cleavage site is chosen such that it covers both the S1/S2 and S2' sites and can be used to screen protease inhibitors against multiple host proteases that cleave the spike protein at different sites to promote viral entry. Once the substrate gets cleaved by the protease, the part of the substrate containing CBD is captured by the cellulose so that only the protein fragment containing the NL sequence remains in the solution and can be further used for luminescent signal detection or the reporter assay.
We report a luminescent substrate protein containing two important cleavages sites (S1/S2 and S2') of SARS-CoV-2 spike protein and highly sensitive nano luciferase for measuring enzyme activity (Figure 1A). Also, the substrate can be easily purified from a bacterial source, making the purification process lesstime consuming and potentially cost effective. The designed substrate is novel for SARS-CoV-2 spike based drug screening as it contains a stretch of spike sequence containing both the S1/S2 and S2' cleavage site, both of which are reported to be important for the viral entry step (Hoffmann and colleagues, Xia et al.7 Tang et al.4). Use of NL protein provides better sensitivity to the assay as it is a bioluminescent protein with a better light emission capacity and enhanced physical and biochemical properties (Hall and colleagues).
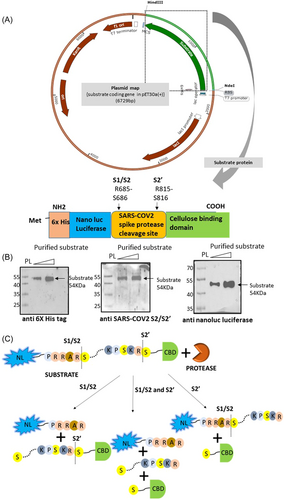
This in vitro assay substrate can potentially be used for screening drugs against host proteases that help in SARS-CoV-2 entry by cleaving at residues lying in the cleavage site between S1 and S2. Candidate inhibitors or drugs that may be screened using the proposed substrate will not essentially compete with the substrate only. There can be two mechanisms: (a) An inhibitor competing with the substrate's binding to the enzyme against which screening is been done and thus disabling binding of the enzyme to the substrate (b) An inhibitor mimicking the substrate binding pocket of the enzyme will bind the substrate and inhibit binding of the enzyme with the substrate thus inhibiting its activity. It can also be used to explore the possible role of any other host protease in the SARS-CoV-2 spike protein priming process. Further, the study highlights a new cleavage point in the SARS-CoV-2 spike between S1/S2 and S2' site (LNRA/LTGI) that has a strong potential to be recognized by host proteases. Currently, the precise SARS-CoV-2 cleavage site for TMPRSS2 is not well characterized. Our study showed that the cleavage sequence LNR/ALTG might be recognized by TMPRSS2. However, further studies are required to confirm this observation.
2 MATERIALS AND METHODS
2.1 Substrate design
The substrate sequence contains His-tag and NanoLuc luciferase sequence (accession ID AIS23666.1) at the N-terminus of the SARS-CoV-2 surface glycoprotein protease cleavage site (accession ID YP_009724390.1; 676th–820th a.a. residues) and the cellulose binding domain (CBD) (accession ID CAA48312.1; 273rd–439th a.a.) at the C terminus. The sequence was codon optimized for bacterial expression and cloned in pET30a vector between NdeI and Hind III restriction sites.
2.2 Substrate purification
Substrate protein expression plasmid was transformed in Nico21 (DE3) cells (New England Biolabs). Transformed colony was added to 2 mL of selection media {LB (Luria-Bertani) + 30 μg/mL Kanamycin (Merck Millipore)}. After 4-6 h incubation at 37°C and 200 rpm shaking condition, the culture was added to a 200 mL selection medium and allowed to grow till OD600 reached 0.4–0.6. The culture was then induced by adding 0.5 mM IPTG (Isopropyl ß-D-1-thiogalactopyranoside) and incubated for 16 h at 15°C under shaking condition. After incubation, the cells were collected by spinning down the culture at 4°C. Cells were re-suspended in 20 mL cell lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM Imidazole pH 8) and sonicated to lyse the cells (30% Amp, 20 s on and 20 s off for 60 min). The soluble (cytoplasmic) and insoluble (inclusion bodies and debris) fractions were separated by centrifuging at 15000g for 30 min at 4°C. The protein was purified from the cytoplasmic fraction by using Ni-NTA column (Qiagen). Briefly, the column was equilibrated by adding 10 mL of binding buffer/cell lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM Imidazole pH 8) and was allowed to flow through by gravity flow. The cytoplasmic fraction was added to the column and after 5 min, the fraction was allowed to pass through. The first washing was performed with 10 mL binding buffer. The second and third washing were performed using 10 mL wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM Imidazole pH 8). The substrate was eluted using the elution buffer (50 mM NaH2PO4, 300 mM NaCl, 200 mM Imidazole pH 8) and dialyzed overnight at 4°C in dialysis buffer (50 mM Tris-HCl, 10% Glycerol, pH 8). The concentration of the purified substrate was checked using BCA assay.
2.3 NanoLuc luciferase assay (NL)
All enzyme-substrate reactions were performed in triplicates and at least three independent times (i.e., at least nine times). Equal volumes of the reaction and 33% w/v matrix slurry (made in the same buffer in which digestion was done) were added in each well of a 96 well plate followed by an incubation in a plate shaker for 15 min at 100 rpm at RT. After incubation, the plate was spun at 2000 g for 7–10 min at RT. Then, 20–30 μL of the supernatant was removed from each well in and placed in separate tubes. The tubes containing the solution were spun shortly to precipitate any extra matrix. From there, 15–20 μL was added to each well followed by the addition of an equal volume of reporter assay reagent (prepared as per the manufacturer's protocol). After adding assay reagent, the plate was incubated for 5 min. Luminescence was recorded for 1 s in Synergy H1 BIOTEK microplate reader.
2.4 Western blot
Western blot was performed to confirm substrate protein expression using antibodies against SARS-CoV2 S2/S2' (GTX135386) (Genetex), Nano Luciferase (N700A) (Promega) and 6x His (GTX115045) (Genetex) antibody. The general protocol followed for gel running, membrane transfer and washing of the blot is same as mentioned below. The antibodies used were as per the samples run.
For detection of cleavage bands by western blot, digestion reaction supernatant was run on SDS-polyacrylamide gel followed by gel transfer at 400 mA for 1.5 h. Membrane was blocked with 5% skimmed milk in TBST for 1 h at RT followed by incubation with anti-NanoLuc Luciferase (N700A) antibody or anti S2/S2' antibody (GTX135386) for 13–15 h at 4°C. Blot was washed in TBST thrice for 5 min followed by incubation with goat anti-mouse IgG2a (NB7516) (NOVUS Biologicals) or goat anti rabbit IgG (NBP175346) (NOVUS Biologicals) for 1 h at RT. After membrane wash, blot was viewed using ECL Clarity (Bio-Rad).
2.5 Substrate digestion with trypsin
2.5.1 Digestion reaction using different amount of trypsin
A total of 500 ng substrate was used with different concentrations of porcine trypsin (Sigma) in 0.1M Tris pH 7.5 in a reaction volume of 50 μL. A total of 500 ng BSA was used as a control. Digestion was done at 37°C for 1 h in water bath followed by SDS-PAGE and Sypro Ruby (Sigma-Aldrich) staining. The same conditions were used for human trypsin (Sigma) as well.
2.5.2 Digestion reaction with varying timepoints
To check the optimum reaction time, 25 ng trypsin was used to digest 500 ng substrate in 0.1 M Tris pH 7.5 in a reaction volume of 50 μL at 37°C and the incubation time was varied from 15–60 min. The reaction volume was loaded in SDS-PAGE gel followed by Sypro Ruby staining.
2.5.3 Inhibition of trypsin-based substrate digestion using inhibitors
A total of 25 ng of trypsin was used to digest 500 ng of substrate at 37°C for 1 h. To check the effect of protease inhibitor (ThermoFisher Scientific) (consisting of AEBSF, aprotinin, bestatin, E-64, leupeptin, and pepstatin A) or trypsin inhibitor (Sigma) on trypsin, different amounts of inhibitor were added to the reaction. Following incubation at 37°C for 1 h, the reactions were used for SDS-PAGE and NL assays.
2.6 Substrate digestion with TMPRSS2
2.6.1 TMPRSS2 based substrate digestion at different timepoints
To check the substrate digestion by active TMPRSS2 (Mybiosource), 250 ng substrate and 250 ng of active TMPRSS2 was used in 1X Tris buffered saline pH 7.5 and the digestion was done at 37°C at different timepoints. Post incubation, the reactions were used for the NL assay.
2.6.2 TMPRSS2 based substrate digestion using different amounts of TMPRSS2
A total of 250 ng of substrate was digested with different amounts of TMPRSS2 in 1X Tris buffered saline pH 7.5 at 37°C for 4 h. Postincubation, the reactions were used for the NL assay.
2.6.3 Inhibition of TMPRSS2 based substrate digestion using camostat mesylate
To check the effect of camostat mesylate on TMPRSS2-mediated digestion, 250 ng of substrate was digested using 250 ng of TMPRSS2 in the presence of camostat mesylate (Sigma) in 1X Tris buffered saline pH 7.5 at 37°C for 2 h. The reactions were further used for the NL assay.
2.7 Substrate digestion with furin
2.7.1 Furin based digestion of substrate using different amounts of the enzyme
Digestion was done in 100 mM HEPES pH 7.5 supplemented with 0.5% Triton X100, 1 mM CaCl2 and 1 mM beta mercaptoethanol at 25°C for 4 h using different units of human Furin (50–800 mU) (Sigma) on 500 ng substrate in a reaction volume of 25 μL. Postincubation, the reaction mixture was run in SDS-PAGE followed by Sypro Ruby staining.
2.7.2 Furin based digestion at different timepoints
A total of 500 ng substrate was digested with 500 mU furin for different timepoints (0.5–4 h) using the same buffer and temperature conditions as mentioned before. The digestion was checked by SDS-PAGE followed by Sypro Ruby staining.
2.7.3 Furin based digestion of substrate in presence of furin inhibitor II
A total of 500 ng of substrate and 500 mU furin were used to perform the substrate digestion in presence of different amounts of furin inhibitor (Sigma) (1–8 ng). After incubation, the reaction was used either for SDS-PAGE followed by Sypro Ruby staining or for NL assay.
2.8 Substrate digestion with cathepsin B
A total of 250 ng substrate was digested with 500 ng human cathepsin B (Sigma) in 1X PBS pH 7.2 and 500 ng substrate was digested with 200 ng cathepsin B in 25 mM MES pH5 for 4 h at 37°C. After the digestion, the reaction mixtures were then run on SDS-PAGE gel followed by western blot analysis.
2.8.1 Cathepsin B based digestion using different amount of enzyme
A total of 100 ng substrate was digested with different amounts of cathepsin B at 37°C for 2 h in 1X PBS pH 7.2 in 25 μL reaction volume. After incubation, NL assay was performed.
2.8.2 Substrate digestion by cathepsin B at different timepoints
A total of 100 ng substrate was digested with 300 ng of cathepsin B in a reaction volume of 25 μL for increasing timepoints (2–4 h) in the same buffer as mentioned before. The reaction mix was further processed for NL assay.
2.8.3 Substrate digestion by cathepsin B in presence of cathepsin B inhibitor II
A total of 100 ng substrate and 300 ng cathepsin B were incubated at 37°C for 2 h in 1X PBS pH7.2 in presence of cathepsin B inhibitor II (Sigma). Post incubation, NL assay was performed.
2.9 Substrate digestion with HAT
2.9.1 Substrate digestion by different amounts of HAT
A total of 500 ng substrate was digested with different amounts of HAT (R and D systems) (5–40 ng) in 50λ of assay buffer (50 mM tris pH 7.5 + 0.05% w/v Brij 35) at 37°C for 3 h followed by SDS-PAGE and Sypro Ruby staining. For NL assay, 250 ng substrate was digested with different amounts of HAT (5–20 ng) at 37°C for 2 h in the same buffer. For immunoblotting, 250 ng substrate was digested with 100 ng HAT in the same buffer at 37°C for 3 h.
2.9.2 HAT based substrate digestion at different timepoints
A total of 250 ng of substrate was mixed with 10 ng of HAT in 25 μL assay buffer and incubated at 37°C for increasing timepoints (1–4 h). Postincubation, NL assay was performed.
2.9.3 HAT based substrate cleavage in presence of camostat mesylate
In 25 μL assay buffer, 250 ng substrate was mixed with 10 ng HAT in presence of different amounts of camostat mesylate (1–4 ng) and incubated at 37°C for 2 h. After incubation, the reaction mix was used for the NL assay.
3 RESULTS AND DISCUSSION
The substrate used for the study was found to be efficiently cleaved by different proteases like trypsin, TMPRSS2, furin, cathepsin B, cathepsin L and HAT that are known to cleave SARS-CoV-2 spike. Western blot analysis for the purified substrate using antibodies against its components confirmed proper translation of the protein with its different domains intact and that no nonspecific protein was purified during the process as the antibodies could not detect any protein other than the substrate in the western blot (Figure 1B). The basic assays used in case of each of the proteases tested on the designed substrate are similar. Inhibitors used were based on the type of enzyme used.
3.1 Specific cleavage of the substrate by trypsin
The substrate used in the assay system was designed such that on cleavage at both the S1/S2 and S2' sites, three bands corresponding to the SARS-CoV-2 spike (∼14 KDa), NanoLuc luciferase (∼21 KDa) and CBD protein (∼19 KDa) would be generated (Figure 1C). Trypsin was found to cleave the substrate at multiple sites including the S1/S2 and S2' (Figure 2, Supporting Information: Figure S1). Trypsin based cleavage shows all these bands as the end product including other intermediate bands arising due to incomplete digestion at only S1/S2 (∼33 KDa) or S2' site (∼35 KDa). Apart from these sites, other monobasic cleavage sites lying between the S1/S2 and S2' sites are also targeted by trypsin and other proteases as protein fragment of around 25–35 KDa were also detected (Figures 2A–D, 3D). When trypsin was added to the substrate in different amounts, the concentration dependent cleavage of the substrate was observed, where trypsin from both the mammalian sources (porcine and human) cleaved the substrate at the same sites (Figure 2A, Supporting Information: Figure S1A). To check for the nonspecific cleavage by trypsin, BSA was used as a control. Porcine trypsin did not digest BSA (Figure 2A) but human trypsin cleaved BSA at sites different from those of substrate (Supporting Information: Figure S1A) owing to its higher activity than that of porcine trypsin as 25 ng porcine trypsin could not digest substrate completely in 1 h reaction time whereas 12.5 ng human trypsin could digest the same amount of substrate completely under the same assay conditions. Further, time dependent cleavage of the substrate was also checked where trypsin could cleave substrate even at 15 min incubation time at 37°C indicating rapid cleavage of the substrate (Figure 2B). To check the effect of trypsin inhibitor in the reaction, it was introduced in different amounts in the reaction mix. Trypsin inhibitor could inhibit trypsin mediated cleavage of the substrate in a concentration dependent manner (Figure 2C, Supporting Information: Figure S2B). The uncleaved substrate band increased in intensity whereas the intensity of the cleavage bands decreased with increasing amount of the inhibitor. Due to the higher specific activity more amount of the inhibitor was required to inhibit human trypsin than trypsin from porcine source. Western blot was performed to identify the trypsin based cleaved bands containing NanoLuc luciferase. Porcine trypsin cleaved the substrate incompletely generating three cleavage bands, 35 KDa (cleavage at S2' site), 25–35 KDa (cleavage between S1/S2 and S2') and 21 KDa (cleavage at S1/S2 site) (Figure 2D) whereas human trypsin digested the substrate completely showing a single band of 21 KDa (Supporting Information: Figure S1C). Western blot was also done using antibody against SARS-CoV-2 spike S2/S2' to confirm the cleavage sites and seven bands could be detected including undigested substrate of 54 KDa (Supporting Information: Figure S4A). Other 6 bands include 33 and 21 KDa bands arising due to cleavage at S1/S2 site, 35 and 19 KDa bands due to S2' site based cleavage and a 14–15 KDa band generated due to cleavage at both the sites (Supporting Information: Figure S4A). Another band of ∼25K Da was also detected due to cleavage between S1/S2 and S2' site which is possible as there are multiple arginine and lysine residues between the two sites. The cleavage site is most likely LNR/ALTG as the fragment size obtained in the western blot analyses using both types of antibodies (anti-NanoLuc luciferase and anti-SARS-CoV-2 spike S2/S2') match with the expected band size of 29 KDa containing NanoLuc luciferase protein and a fragment of 25 KDa (Figure 2D, Supporting Information: Figure S4A). However, further studies like mass spectrometry and mutagenesis may be required to confirm the cleavage site.
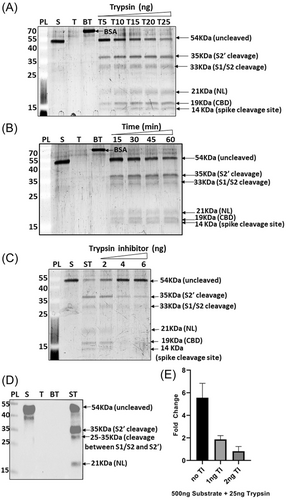
To check the fold change in luminescence due to substrate digestion, the NL assay system was used where the cleaved substrate showed a 5.5 fold change in luminescence as compared to the undigested substrate. The fold change further reduced to 1.9 and 0.8 when trypsin inhibitor (TI) was introduced in the reaction in two different amounts; 1 and 2 ng, respectively (Figure 2E). When human trypsin was used to digest the same amount of substrate under the same experimental condition, a 27.4 fold change in luminescence was detected which reduced to 12.7 and 7.1 fold change when 10 and 20 ng of TI was added to the reaction, respectively (Supporting Information: Figure S1D). Hence, human trypsin was much more efficient in cleaving the substrate than the porcine trypsin. Human trypsin was used for the study to better correlate with human airway trypsin like protease (HAT) that might have a significant role in mediating SARS-CoV-2 entry in human lung epithelium7 and Kim et al.9). Therefore, HAT may possibly cleave substrate similar to that of human trypsin by recognizing both S1/S2 and S2' site along with other intermediate sites having arginine and lysine residues.
We also included protease inhibitor cocktail (PIC) in the experiment to study its effect on substrate cleavage by trypsin (Supporting Information: Figure S2). When used in different amounts, PIC could inhibit the substrate cleavage (Supporting Information: Figure S2A) where the cleavage band intensity decreased with increasing amounts of protease inhibitor. Similarly, in the NL assay the fold change reduced gradually with increasing amounts of PIC (Supporting Information: Figure S2B). The decrease in fold change due to PIC was lesser than that of TI. This could be due to the fact that PIC is a combination of multiple inhibitors out of which some might not be working as trypsin inhibitors. TI being specific to trypsin could be inhibiting digestion by trypsin even in small amounts.
3.2 Cleavage of substrate by TMPRSS2, furin and cathepsins
Different host proteases like trypsin and trypsin like proteases, TMPRSS2, furin and cathepsin B are known to cleave SARS-CoV-2 spike protein between S1 and S2 domains. Our substrate contains the entire cleavage site between S1/S2 and thus the proposed assay system should be potentially useful in screening drugs against multiple proteases cleaving the S1/S2 cleavage site. Thus, after primary validation of the assay system using trypsin, we checked if the substrate can get cleaved by other host proteases that are known to cleave SARS-CoV-2 spike as well. TMPRSS2, furin, cathepsin B, cathepsin L and HAT were used for further validation of the substrate cleavage.
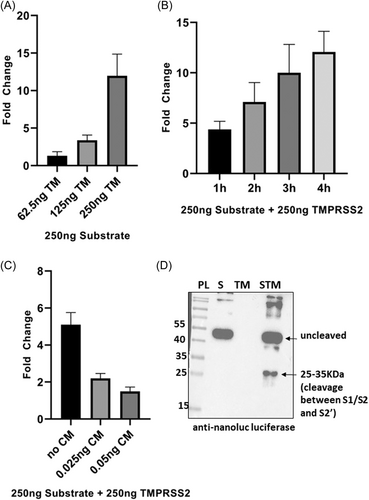
TMPRSS2 is one of the important host proteases that is known to help in SARS-CoV-2 entry by cleaving spike protein although the precise cleavage site of the enzyme is not well documented. Recent work by Bestle et al.16 speculate SARS-CoV-2 spike S2' site to be the most likely cleavage point for TMPRSS2.16 In our assay system, TMPRSS2 could cleave the SARS-CoV-2 spike between the S1/S2 and S2' cleavage site, thereby releasing a 25–35 KDa fragment from the substrate that contains the NL protein (Figure 3D). The same fragment could also be detected when substrate was cleaved by porcine trypsin and cathepsin B (Figures 2D, and 5A,B) indicating that this cleavage point is common for all the three enzymes. Also, S2' site in SARS-CoV-2 spike possesses both the basic residues (arginine and lysine) together, hence creating a dibasic site (PSKPSKR/SF) which might not be recognized by TMPRSS2 as it is known to cleave after a monobasic residue (Hoffman and colleagues, Bestle et al.16). Work on MERS-CoV spike showed that the S2' site is involved in TMPRSS2 based protein priming based on the fact that S2' site of MERS-CoV spike contains two arginine residues separate from each other that makes it a monobasic site and hence suitable for cleavage by this enzyme.15
We checked substrate cleavage by TMPRSS2 using different amounts of the enzyme. The fold change in luminescence was found to be 1.3, 3.4, and 12 when 62.5, 125, and 250 ng TMPRSS2 was used to digest 250 ng substrate, respectively (Figure 3A). Also, there was a significant increase in fold change when incubation time was increased from 1 to 4 h (Figure 3B). Addition of TMPRSS2 inhibitor, camostat mesylate (CM) in the reaction lead to a significant decrease in the fold change (Figure 3C). Use of active TMPRSS2 purified from mammalian source showed increase in fold change from 3.4 to 5.3 when incubation time was increased from 4 h to 6 h, respectively (Supporting Information: Figure S3A). Mammalian purified active TMPRSS2 resembles human TMPRSS2 based on posttranslational modifications and hence the substrate cleavage by mammalian TMPRSS2 is indicative of similar activity of human TMPRSS2.
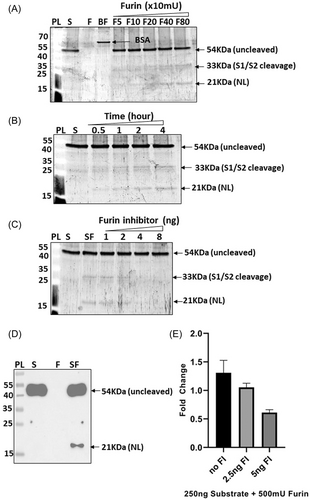
Furin is known to cleave SARS-CoV-2 spike sequence at S1/S2 site.2, 3 As shown in Figure 4A, 500 ng substrate was digested with different amounts of furin (50–800 mU) and a gradual increase in intensity of the two cleavage bands (33 and 21 KDa) corresponding to S1/S2 cleavage were observed. When the reaction was done using a fixed amount of substrate and furin and the incubation time was increased gradually, the cleavage bands were found to increase parallelly with the incubation time. Furin inhibitor could inhibit the digestion efficiently (Figure 4C) indicating that the substrate cleavage was furin specific. To further confirm the cleavage site, western blot analysis was done using anti NL and anti-SARS-CoV-2 spike S2/S2' antibodies (Figure 4D and Supporting Information: Figure S4B). When anti SARS-CoV-2 spike S2/S2' antibody was used, two fragments, 33 and 21 KDa containing the SARS-CoV-2 spike sequence were detected, hence confirming the S1/S2 site-based cleavage by furin (Supporting Information: Figure S4B) as shown by other groups.2, 3 Western blot analysis using anti NL antibody showed a band of 21 KDa, thereby correlating with the previous data (Figure 4D). We further checked the fold change in luminescent signal due to furin based substrate cleavage and a 1.3-fold change was observed when substrate was treated with furin as compared to untreated substrate (Figure 4E). Introduction of furin inhibitor in different amounts (2.5 and 5 ng) in the reaction reduced the fold change to 1.05 and 0.61, respectively (Figure 4E). The fold change observed due to furin based cleavage was comparatively lesser that that of other proteases likely due to the buffer used for the reaction. The buffer components might have an influence over the bioluminescence activity of the substrate owing to the fact that the cleaved substrate bands were clearly visible in the SDS-PAGE gels (Figure 4A–C) but was less detectable in NL assay.
Cathepsin B, which is also a well-known host protease that mediates SARS-CoV-2 entry was also included in our assay for further confirmation. The substrate was found to be cleaved by cathepsin B at two different pH conditions: 5 (lysosomal pH) and 7.2 (cytosolic pH) as the enzyme is located at both the cellular compartments.17 As per the western blot analysis, the substrate was susceptible to cathepsin B cleavage at both the pH conditions but the cleavage sites differed for the two conditions such that at an acidic pH two cleaved bands of size ∼35 and ∼30 KDa were detected whereas at cytosolic pH a band ∼22 KDa was observed (Figure 5A–C). Such pH specific cleavage site preference of cathepsin B has been previously reported which arises due to the ability of the enzyme to act both as an endopeptidase and dipeptidyl carboxypeptidase depending on the pH.17 Bollavaram et al.18 identified two potential cleavage sites of cathepsin B in SARS-CoV-2 spike: LNRA/LTGI (in S2 region) and AYTM/SLGA (near S1/S2 site). As per our analysis, at cytosolic pH, the enzyme is possibly cleaving substrate at AYTM/SLGA site thereby generating a ∼22 KDa band (Figure 5C) whereas at lysosomal pH, LNRA/LTGI site is preferred for cleavage as a ∼30 KDa band appeared along with a ̴35 KDa which is likely due to cleavage near the S2' site as seen in Figure 5A,B. We further studied the enzyme cleavage efficiency by using NL assay where the enzyme amount was varied keeping other conditions same and a six fold change was obtained when the substrate was digested with 300 ng of cathepsin B (Figure 5D). The fold change reduced to 5.4and 1.9 when cathepsin B was reduced to 200 and 100 ng, respectively (Figure 5D). With increasing Incubation time there was a significant increase in the luminescent signal when a fixed amount of substrate and enzyme was used (Figure 5E). The fold change was observed to be 5.3, 10.5 for 2 and 4 h, respectively. Introduction of cathepsin B inhibitor reduced the fold change to 3.4 and 2.6 when 0.125 and 0.25 ng of the inhibitor were used respectively (Figure 5F).
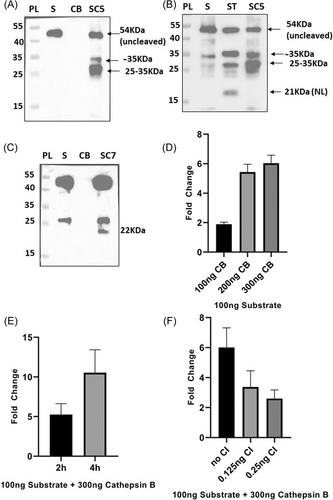
Cathepsin L had been shown to be important in SARS-CoV2 entry.10 Hence, we also checked the activity of Cathepsin L on the substrate by using different enzyme concentrations. When substrate was treated with increasing amounts of cathepsin L (12.5–50 ng), the fold change of substrate cleavage with respect to no enzyme control increased from 4.3 to 7.2 (Supporting Information: Figure S5).
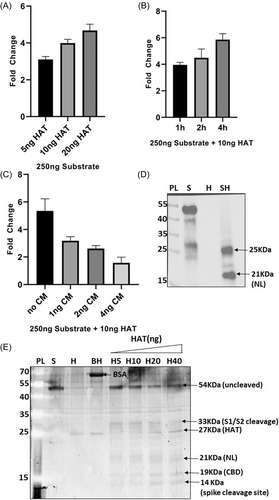
HAT (also known as TMPRSS11D) was further used in the cleavage assay. When the enzyme amount was varied while maintaining the amount of substrate constant, a gradual increase in the fold change of luciferase activity was observed in NL assay (Figure 6A). Also, the luminescent signal was found to increase with increasing incubation time (1–4 h) (Figure 6B). When camostat mesylate (a potent serine protease inhibitor) was introduced in the reaction {HAT, being one of the members of Type II serine proteases (TTSPs) family, closely resembles TMPRSS2 structurally and functionally}, the fold change in luminescent signal was found to gradually decrease with increasing amount of the inhibitor (1–4 ng) (Figure 6C). To check for the cleavage sites, SDS-PAGE gel analysis was performed in which three prominent bands (NL, CBD and spike cleavage sites) were obtained indicating cleavage at both S1/S2 and S2' site (Figure 6E). Immunoblot results confirm the band for NL (21 KDa) with an additional band of ∼25 KDa (likely due to cleavage at another site between S1/S2 and S2' regions) (Figure 6D).
The results as discussed above for each enzyme has been summarized in Table 1.
| Protease | Reaction conditions | Site of cleavage | Fold change after cleavage | Protease inhibitor used | Fold change after addition of protease inhibitor |
|---|---|---|---|---|---|
| Trypsin (porcine source) | Digestion at 37°C for 1 h in in 0.1 M Tris pH 7.5 | S1/S2; S2'; between S1/S2 and S2' | 5.6 | Trypsin inhibitor from glycine max | 1 ng = 1.9 2 ng = 0.8 |
| Trypsin (porcine source) | Digestion at 37°C for 1 h in in 0.1 M Tris pH 7.5 | S1/S2; S2'; between S1/S2 and S2' | 8.9 | Protease inhibitor cocktail | 2.5 μL = 7 10 μL = 4.5 |
| Trypsin (human source) | Digestion at 37°C for 1 h in in 0.1 M Tris pH 7.5 | S1/S2; S2' | 27.4 | Trypsin inhibitor from glycine max | 10 ng = 12.7 20 ng = 7.1 |
| Human airway trypsin like protease (HAT) | Digestion at 37°C for 2 h in assay buffer(50 mM Tris pH 7.5 + 0.05% w/v Brij 35) | S1/S2; S2’; between S1/S2 and S2' | 5.3 | Camostat mesylate | 1 ng = 3.2 4 ng = 1.6 |
| TMPRSS2 | Digestion at 37°C for 2hrs in 1X Tris buffered saline pH 7.5 | Between S1/S2 and S2' | 5.1 | Camostat mesylate | 0.025 ng = 2.2 0.05 ng = 1.5 |
| Furin | Digestion at 25°C for 5 h in 100 mM HEPES pH7.5 + 0.5% TritonX100 1 mM CaCl2 and 1 mM beta mercapto ethanol | S1/S2 | 1.3 | Furin inhibitor | 2.5 ng = 1.1 5 ng = 0.6 |
| Cathepsin B | Digestion at 37°C for 2 h in 1X PBS pH 7.2 | Between S1/S2 and S2' | 6 | Cathepsin B inhibitor | 0.125 ng = 3.4 0.25 ng = 2.6 |
4 SUMMARY
The substrate reported in this manuscript has been validated for cleavage ability using multiple host proteases involved in the viral ingress. In the NL assay, a significant fold change was observed when the substrate was subjected to digestion with enzymes. The enzymatic digestion decreased when inhibitors were introduced in the reaction. Furthermore, SDS-PAGE and immunoblotting results helped to identify the cleavage sites for individual enzymes. Specifically, we used two important proteases: HAT and TMPRSS2 for our study which are not well studied in context to cleavage site in the spike protein. Using this assay system, we identified a few cleavage positions (LNRA/LTGI and AYTM/SLGA) lying between S1/S2 and S2' sites that may be involved in the priming process. LNRA/LTGI site could be a common site of recognition by proteases like TMPRSS2, cathepsin B and trypsin. The study highlights the multipurpose use of the designed substrate for screening inhibitors against multiple enzymes, studying cleavage sites for enzymes, identifying potential host proteases involved in spike cleavage and to study the interaction between different enzymes when introduced together.
AUTHOR CONTRIBUTIONS
Feroza Begum performed all experiments, analyzed data and wrote the manuscript. Amit K. Srivastava helped with the proofreading. Prem Prakash Tripathi helped with data analysis and proofreading. Upasana Ray conceptualized and designed the assay system, analyzed data and wrote the manuscript.
ACKNOWLEDGMENTS
The work was partly funded by the Council of Scientific and Industrial Research (CSIR) under project code MLP-131 LRF and also by other intramural CSIR-IICB laboratory funds. We thank CSIR and AcSIR for supporting Ms Feroza Begum.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Data will be available on request from the authors.




