High detection rate of viral pathogens in nasal discharge in children aged 0 till 5 years
Elandri Fourie and Yara E. E. Sijm contributed equally to this study.
Abstract
Respiratory tract infections (RTI) in children remain a cause of disease burden worldwide. Nasopharyngeal (NP) & oropharyngeal (OP) swabs are used for respiratory pathogen detection, but hold disadvantages particularly for children, highlighting the importance and preference for a child friendly detection method. We aimed to evaluate the performance and tolerability of a rhinorrhea swab (RS) in detecting viral pathogens when compared to a combined OP(/NP) or mid-turbinate (MT) nasal swab. This study was conducted between September 2021 and July 2022 in the Netherlands. Children aged 0−5 years, with an upper RTI and nasal discharge, were included and received a combined swab and a RS. Multiplex polymerase chain reaction (PCR) and severe acute respiratory syndrome coronavirus-2 PCR were used for viral pathogen detection. Tolerability was evaluated with a questionnaire and visual analog scale (VAS) scores. During 11 months 88 children were included, with a median age of 1.00 year [interquartile range 0.00−3.00]. In total 122 viral pathogens were detected in 81 children (92%). Sensitivity and specificity of the RS compared to a combined swab were respectively 97% (95% confidence interval [CI] 91%−100%) and 78% (95% CI 45%−94%). Rhinorrhea samples detected more pathogens than the (combined) nasal samples, 112 versus 108 respectively. Median VAS scores were significantly lower for the RS in both children (2 vs. 6) and their parents (0 vs. 5). A RS can therefore just as effectively/reliably detect viral pathogens as the combined swab in young children and is better tolerated by both children and their parents/caregivers.
Abbreviations
-
- MTS
-
- nasal mid-turbinate swab
-
- NPS
-
- nasopharyngeal swab
-
- NS
-
- nasal swab
-
- OPS
-
- oropharyngeal swab
-
- PCR
-
- polymerase chain reaction
-
- RS
-
- rhinorrhea swab
-
- SARS-CoV-2
-
- severe acute respiratory syndrome coronavirus-2
-
- URTI
-
- upper respiratory tract infection
-
- VAS
-
- visual analog scale
1 BACKGROUND
Upper respiratory tract infections (URTIs) remain an important cause of disease burden worldwide, with the highest incidence seen among children under the age of 5 years.1 These infections are generally characterized by throat irritation, coughing, a stuffy or a runny nose, sneezing, headache, fever, and general malaise.2 Our respiratory tract microbiome can be considered as the gatekeeper for respiratory health, yet when microbial imbalance and colonization by respiratory microorganisms takes place, infections can be the aftermath.3 Clinical symptoms caused by these various pathogens are typically overlapping, which implies that laboratory testing is required for definitive diagnosis.4 Identification of the causative viral pathogens may contribute to individual patient care, appropriate (hospital) infection control measures and epidemiological surveillance.4, 5
Over the years, the development of more sensitive laboratory tests has allowed for a less invasive sampling procedure to be developed for respiratory pathogen detection.6 Nowadays, molecular diagnostics are widely applied and the nasopharyngeal swab (NPS) is frequently considered to be the gold standard for viral pathogen detection.7, 8 However, the NPS is an invasive sampling method and known to cause discomfort in both young and old patients, with children in particular being less willing to endure this unfriendly method.
During an URTI, secretions from the nasopharynx, nasal cavity, and sinuses, are discharged from the nose. In theory, pathogens colonizing these different niches should be detectable in these secretions.9 Previous studies have compared different sampling methods, but the identification of pathogens in nasal discharge is a less studied topic.10 Two studies examining the detection of bacteria from nasal discharge, particularly collected from a tissue, showed promising results. Leach et al. compared nasal swabs (NSs) with swabs from nasal discharge in a tissue and found a high sensitivity detection rate of 94% for pneumococci from nasal discharge in children with visible secretions.11 Furthermore, Van den Bergh et al. sampled 66 children by using NPSs, NSs and nose blowing or wiping into a tissue and found that bacterial pathogens were reliably detected by these different sampling techniques.9 One study performed by She et al. compared NPS with nasal mucus in tissues and found that viruses in nasal mucus were detected in 73% (11/15) of the positive cases.12 In addition, Blaschke et al. evaluated the utility of both anterior NSs and tissues in comparison to NP aspirates and found a sensitivity for tissues ranging from 33% to 84% for individual viruses.13 However, none of these studies examined the detection of viruses directly from nasal discharge.
Respiratory viruses circulate throughout the year, peak at certain time points and occasionally cause major outbreaks with serious impact on global health.14 The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic has underlined the importance of rapid identification and epidemiologic tracing of viral pathogens.15 In the Netherlands, the combined nasal mid-turbinate swab (MTS) and oropharyngeal swab (OPS) was recommended in children under 13 years of age for, among others, the diagnosis of SARS-CoV-2.16 Since children have been tested more frequently during the SARS-CoV-2 pandemic, it would be preferred to have the availability over a more child-friendly sampling method. A noninvasive method might be better for their overall experience, could reduce resistance toward testing and may lead to higher quality samples due to an effortless execution. Alternative noninvasive methods such as saliva swabs or collectors have already been proven to have a high concordance rate of 90% when compared to NPS.17 However, this method requires special tubes or sponges for saliva collection, which can be expensive, difficult and time-consuming for laboratory testing.
As mentioned earlier, up to date no study has examined the detection of viral pathogens directly from nasal discharge, which lead us to the specific opportunity to evaluate this method in young children.
The primary objective of this study was to compare the performance of a rhinorrhea swab (RS) to that of the gold standard, the (combined) NP/MT nasal (and OP) swab, in detecting respiratory pathogens in young children using multiplex polymerase chain reaction (PCR) and SARS-CoV-2 reverse transcriptase PCR. The secondary objective was to evaluate the tolerability of the different testing methods using a Visual Analog Scale (VAS) score. It is hypothesized that an RS can reliably detect respiratory pathogens as an alternative method to that of the standard care and that this RS will be much better tolerated.
2 METHODS
2.1 Setting and design
For this prospective observational study participants were randomly recruited from September 2021 to July 2022. Recruitment took place at two centers: at the emergency room and the pediatric ward of the Spaarne Gasthuis (hereafter sometimes referred to as “SG”) hospital located in Haarlem (The Netherlands) and at the Public Health Service corona testing facility at Schiphol (hereafter sometimes referred to as “GGD”).
2.2 Participants
Children aged between 0 and 5 years, presenting at one of the centers with symptoms/signs of an URTI, characterized by at least nasal discharge, and an indication for a routine nasal swab, were eligible for participation. Infants were excluded if they had received antiviral therapy in the last 2 weeks before recruitment, were too sick to participate as objectified by a medical health professional, had craniofacial abnormalities and if their parents or caregivers were unable to give informed consent due to insufficient understanding of the Dutch or English language. Among others, gender, age, results of both sampling methods and duration of admission were recorded for each participant. The code of conduct relating to expressions of objection by minors participating in medical research was adhered to.
2.3 Materials and data collection
Two samples were collected for each individual. The eSwabTM 490CE (COPAN), a swab and a tube with 1 ml liquid Amies preservation medium, was used for collection of the rhinorrhea sample. Using the eSwab nasal discharge was collected at the beginning of the nasal entrance or on the upper lip. The eSwab was taken directly from the nasal discharge or, if this was preferred by the parent(s) or caregiver(s) or the child, from a tissue paper after wiping their nose. When nasal discharge was not present, the child was asked to blow their nose and, when secretion was visible, the sample was taken from the tissue paper. WEPA Satino® prestige 2-layer tissues were used as tissue paper for rhinorrhea collection.
A routine swab was then performed according to standard procedures, generally a (combined) MT nasal (and OP) swab, hereafter termed nasal swab. Nasal swab was chosen since this type of swab was certainly performed in all children.16, 18-20 Combined NP and OP swabs were not collected during this study, due to the change in sampling preference during the COVID-19 pandemic.21 The iClean® Specimen Collection Flocked swab was used for routine sample collection and the Remel MicroTestTM M4RT® Transport 3.0 ml tube was used as transport medium. Disposable gloves, face masks and aprons were worn during both sampling procedures.
Ideally the RS and NS were to be taken in a fixed order, immediately after one another, with the RS sample being taken first. However, children did not always have nasal discharge at the exact moment that the routine swab was performed, nonetheless all samples were collected with a maximum of 24 h between two samples.
In addition, a questionnaire was used to evaluate the presenting symptoms of the child's respiratory infection such as the duration of symptoms, the type of symptoms, the color of rhinorrhea and the amount of rhinorrhea. To measure tolerability across both sampling methods, for both children and their parent(s) or caregiver(s), four VAS scores were noted.
2.4 Laboratory analysis
Both samples were stored and transported to the Regional Public Health Laboratory Kennemerland laboratory according to standard procedures at room temperature. Both samples were analyzed within 48 h using multiplex ligation-dependent probe amplification for the presence of 4 bacterial and 18 viral pathogens: bordetella pertussis; chlamydia pneumoniae; legionella pneumophila; mycoplasma pneumoniae; adenovirus; human bocavirus; coronavirus OC43, 229E, and NL63/HKU1; influenza virus A, B, and A(H1N1)pdm09; human metapneumovirus; parainfluenza virus 1, 2, 3, and 4; rhinovirus/enterovirus; RSV A and B (RespiFinder® 2Smart; PathoFinder). In addition, the samples were analyzed for the presence of SARS-CoV-2 using reverse transcriptase PCR. Amplification products were detected with the Roche LightCycler® 480.
2.5 Main outcome measures and statistical analysis
The performance of an RS was compared to that of a (combined) nasal MT (and OP) swab for detecting viral pathogens using multiplex PCR. Data analyses were performed using the statistical software package SPSS version 28. Means with standard deviation (SD) were calculated for normally distributed continuous variables and medians with interquartile range (IQR) for skewed distributed continuous variables. Proportions were calculated for dichotomous and categorical variables. A nonparametric independent sample t-test (Mann−Whitney U test) was used to evaluate the differences in age and duration of symptoms between the two groups. In addition, the chi-square test or the fisher's exact test was used to calculate differences in other characteristics between the two groups.
The primary outcome measures were the sensitivity and specificity of the multiplex PCR and SARS-CoV-2 reverse transcriptase PCR results for both the RS and NS, when using the NS as the gold standard. This was calculated using 2 × 2 contingency tables. The McNemar test was used to test for differences in the proportions. In addition, concordance rates were calculated and confidence intervals (CI) were calculated using epitools.22 Wilson CIs were calculated if the proportion ranged from [5% to 95%] and otherwise Clopper−Pearson exact intervals were calculated. The secondary outcome measures were the tolerability, calculated as the differences in VAS score across both sampling methods, analyzed using the nonparametric Wilcoxon Signed Rank test, assuming a non-normal distribution. The VAS score measurement level was considered as a ratio since absolute differences were evaluated.23 A p value below 0.05 was considered statistically significant.
3 RESULTS
3.1 Participant characteristics
A total of 91 children were recruited during a period of 11 months. Three children were not included in the study due to a lack of a reference/NS sample, leading to 88 children being included. The majority of these children (n = 68) were recruited at the emergency department and the pediatric ward of the Spaarne Gasthuis in Haarlem. Twenty children were recruited at the Public Health Service corona testing facility at Schiphol. All children were included in the analysis, including one child with craniofacial abnormality, as there was no contraindication for routine sampling and therefore also not for the RS.
The median age of participants was 1.00 years [IQR 0.00−3.00], ranging from 0 to 5 years, with the majority of children being girls (53%). Characteristics of the participants are presented in Table 1. No significant differences were found between the groups for gender (p = 0.871) or duration of symptoms (p = 0.075), however, there was a significant difference in age (p = 0.003). Median duration of symptoms at the time of presentation was 5 days [IQR 3.00−6.00] with the most commonly reported complaints being coughing (94%), fever (66%), shortness of breath (65%), fatigue (65%) and sneezing (65%). The majority of the reported colors and amounts of rhinorrhea were respectively bright (61%) and mediocre (41%).
| Total (n = 88) | SG (n = 68) | GGD (n = 20) | p Value | |
|---|---|---|---|---|
| Demographic characteristics | ||||
| Median age in years [IQR] | 1.00 [0.00−3.00] | 1.00 [0.00−2.00] | 3.00 [0.25−5.00] | 0.003c |
| Age group—n (%) | <0.001d | |||
| 0 years | 33 (38%) | 28 (41%) | 5 (25%) | |
| 1 year | 21 (24%) | 20 (29%) | 1 (5%) | |
| 2 years | 10 (11%) | 10 (15%) | 0 (0%) | |
| 3 years | 10 (11%) | 4 (6%) | 6 (30%) | |
| 4 years | 4 (5%) | 3 (4%) | 1 (5%) | |
| 5 years | 10 (11%) | 3 (4%) | 7 (35%) | |
| Sex—n (%) | 0.871d | |||
| Male | 41 (47%) | 32 (47%) | 9 (45%) | |
| Female | 47 (53%) | 36 (53%) | 11 (55%) | |
| Clinical characteristics | ||||
| Median duration of symptoms in days [IQR] | 5.00 [3.00−6.00] | 5.00 [4.00−7.00] | 3.5 [2.00−5.00] | 0.075c |
| Symptoms—n (%)a | ||||
| Coughing | 83 (94%) | 65 (96%) | 18 (90%) | 0.318d |
| Earache | 7 (8%) | 7 (10%) | 0 (0%) | 0.343d |
| Sneezing | 57 (65%) | 44 (65%) | 13 (65%) | 0.981d |
| Headache | 10 (11%) | 3 (4%) | 7 (35%) | <0.001d |
| Diarrhea | 17 (19%) | 14 (21%) | 3 (15%) | 0.752d |
| Vomiting | 19 (22%) | 17 (25%) | 2 (10%) | 0.220d |
| Hoarseness | 25 (28%) | 23 (34%) | 2 (10%) | 0.038d |
| A sore throat/pain when swallowing | 24 (27%) | 19 (28%) | 5 (25%) | 0.795d |
| Fever | 58 (66%) | 43 (63%) | 15 (75%) | 0.329d |
| Shortness of breath | 57 (65%) | 54 (79%) | 3 (15%) | <0.001d |
| Fatigue | 57 (65%) | 47 (69%) | 10 (50%) | 0.116d |
| Color of rhinorrhea—n (%)a | 0.670d | |||
| Bright | 54 (61%) | 38 (56%) | 16 (80%) | |
| Green | 13 (15%) | 11 (16%) | 2 (10%) | |
| Yellow | 9 (10%) | 8 (12%) | 1 (5%) | |
| Otherb | 12 (14%) | 11 (16%) | 1 (5%) | |
| Amount of rhinorrhea—n (%)a | 0.465d | |||
| A lot | 30 (34%) | 23 (34%) | 7 (35%) | |
| Mediocre | 36 (41%) | 29 (43%) | 7 (35%) | |
| Little | 18 (21%) | 12 (18%) | 6 (30%) | |
| A lot/mediocre | 4 (5%) | 4 (6%) | 0 (0%) | |
| Admission—n (%) | <0.001d | |||
| Yes | 59 (67%) | 59 (87%) | 0 (0%) | |
| No | 29 (33%) | 9 (13%) | 20 (100%) | |
| Duration of admission in days [IQR] | 2.00 [0.00−3.00] | 2.00 [0.00−3.00] | NA | NA |
- Abbreviations: IQR, interquartile range; n, number; NA, not applicable.
- a Sometimes more than one option was selected on the questionnaire. These combinations are presented. Based on subjective observation by parent/caregivers.
- b Other includes any combinations of bright/green/yellow/red reported.
- c Mann−Whitney U test.
- d Chi-square test or Fisher's exact test where appropriate.
When comparing the two centers, some differences in reported complaints were identified. The children recruited at the GGD reported more headaches (35%) and fever (75%) and less earaches (0%) compared to children at the SG center, whose symptoms were more frequently shortness of breath (79%), fatigue (69%), and hoarseness (34%).
Paired swab samples were performed in all included children. MTSs (18%) or combined MTSs and OPSs (49%) were performed in most cases for sample collection, however in 33% of the cases the exact method for sampling of NS is unknown. RS were mainly taken directly from nasal discharge (85%). If nasal discharge was not present at the time of sample collection, but the child was able to blow their nose, the sample was taken from tissue paper (15%). Samples were taken from visible snot or visible moist on the tissue paper and only dry tissues were discarded. Eventually, RSs were taken more frequently after the routine sample collection (70%). An overview of the order and methods of sample collection are described in Table 2. Median time between both sampling methods was 60 min [IQR 1.00−935.00], ranging from 1 min to a maximum of 24 h. Both samples were collected within 1 h in 47 (53%) of the participants and within 6 h in 56 (64%) of the participants.
| Total (n = 88) | SG (n = 68) | GGD (n = 20) | p Valuea | |
|---|---|---|---|---|
| Nasal swab | <0.001 | |||
| Mid-turbinate | 16 (18%) | 16 (24%) | 0 (0%) | |
| Mid-turbinate and oropharyngeal | 43 (49%) | 23 (34%) | 20 (100%) | |
| Unknown | 29 (33%) | 29 (43%) | 0 (0%) | |
| Rhinorrhea swab | 0.481 | |||
| Beginning of nasal entrance or on the upper lip | 75 (85%) | 59 (87%) | 16 (80%) | |
| Tissue paper after blowing the nose | 13 (15%) | 9 (13%) | 4 (20%) | |
| Collection order | 0.005 | |||
| Rhinorrhea swab first | 26 (30%) | 15 (22%) | 11 (55%) | |
- a χ2 test or Fisher's exact test where appropriate.
3.2 Primary outcome measures
Eighty-one (92%) of the 88 participating children were positive for one or more viruses. The RS and NS were both positive for at least one pathogen in 79 (90%) of these children (McNemar's test p = 1.000). Codetections were found in 32 (36%) children, with two and three viruses being simultaneously detected in respectively 19 (22%) and 7 (8%) of the rhinorrhea samples and 19 (22%) and 5 (6%) of the nasal samples. One paired swab sample showed a codetection with four viruses within one participant, namely for rhinovirus, adenovirus, human metapneumovirus, and human bocavirus. Results of the samples are presented in Table 3.
| Positivea | Positive for one virus | Positive for two viruses | Positive for three viruses | Positive for four viruses | Negative | Total | |
|---|---|---|---|---|---|---|---|
| Rhinorrhea sample | 79 (90%) | 53 (60%) | 19 (22%) | 7 (8%) | 0 (0%) | 9 (10%) | 88 (100%) |
| Nasal sample | 79 (90%) | 55 (63%) | 19 (22%) | 5 (6%) | 0 (0%) | 9 (10%) | 88 (100%) |
| Either sampleb | 81 (92%) | 49 (56%) | 24 (27%) | 7 (8%) | 1 (1%) | 7 (8%)c | 88 (100%) |
- a Samples were considered positive if at least one pathogen was detected.
- b Either sample means that a virus was only counted once, even if it was detected in both sample types.
- c Here “either sample” means that both the samples were negative for pathogens.
In total, 122 viral pathogens were detected by either sampling method, revealing 15 different viruses. No bacteria were detected with either sampling method. Rhinovirus was the most frequently detected virus, as it was detected by either sampling method in 41 (47%) children. Overall, the RS detected 112 (92%) viral pathogens, compared to 108 (89%) viral pathogens detected by the NS. Adenovirus, human bocavirus and respiratory syncytial virus B were more frequently detected by the RS and human metapneumovirus, rhinovirus and respiratory syncytial virus A were more frequently detected by the NS. The number of pathogens detected by the different sampling methods is presented in Figure 1. Most viruses detected were found between October 2021 and February 2022 and involved viruses such as adenovirus, human bocavirus, human metapneumovirus, rhinovirus and RSV-A & RSV-B. Some viruses were also detected in April, June and July, with the majority being adenovirus, influenza-A, influenza-A(H1N1), parainfluenza-3, rhinovirus, and some RSV-A.
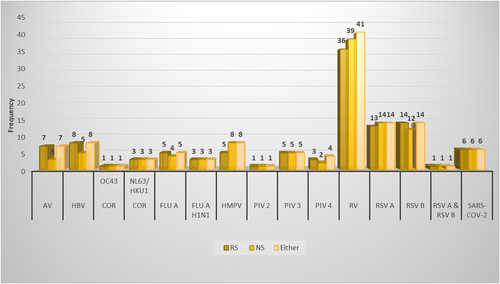
RSs were positive for at least one viral pathogen in 77 of the 79 children with a positive NS, which resulted in a sensitivity for the RS of 97% (95% CI 91%−100%). RSs were negative in 7 of the 9 children with a negative NS, which resulted in a specificity of 78% (95% CI 45%−94%). The results of the RS compared to the reference standard, the NS or (combined) MT nasal (and OP) swab, are presented in Table 4.
| Nasal samplea | |||
|---|---|---|---|
| Rhinorrhea Sample | Positive | Negative | Total |
| Positive | 77 | 2 | 79 |
| Negative | 2 | 7 | 9 |
| Total | 79 | 9 | 88 |
- Note: True positive result: both the rhinorrhea sample and nasal sample are positive for detection of at least one pathogen; true negative result: both the rhinorrhea sample and nasal sample are negative for pathogen detection; false positive result: the rhinorrhea sample is positive, and the nasal sample is negative for detection of at least one pathogen; false negative result: the rhinorrhea sample is negative, and the nasal sample is positive for detection of at least one pathogen. Sensitivity = 77/79 = 97% (95% CI 91%−100%); specificity = 7/9 = 78% (95% CI 45%−94%).
- Abbreviations: CI, confidence interval.
- a The reference sample is the nasal sample, which was a (combined) mid-turbinate nasal (and oropharyngeal) sample.
Concordance between both samples for the detection of at least one pathogen was 95% (95% CI 89%−99%). Concordance between both samples for a complete match was 78% (95% CI 69%−86%). The concordance rate is presented in Table 5 (further details can be found in the Supporting Information File: Table 5) and an overview of the reported outcome measures is presented in Table 6. In total the RS missed 10 viruses that were detected by the NS. The RS missed rhinovirus five times, human metapneumovirus three times, parainfluenza-4 once, and RSV-A once. Yet when only using the RS as diagnostic sample, 14 additional viruses were found, namely adenovirus in four samples, human bocavirus in three samples, RSV-B in two samples, rhinovirus in two samples, parainfluenza-4 in two samples and influenza-A in one sample. Concordance for a complete match between the NS and rhinorrhea samples taken from tissues was 85% (11/13 samples) and for the NS and rhinorrhea samples it was 77% (58/75 samples).
| Parameter | Number (%) |
|---|---|
| Concordanta paired swabs | 69 (78%) |
| No virus identified | 7 (8%) |
| Single virus identified | 46 (52%) |
| Two viruses identified | 12 (14%) |
| Three viruses identified | 4 (5%) |
| Disconcordantb paired samples | 19 (22%) |
- a Concordant was defined as a complete match of the rhinorrhea sample and the nasal sample.
- b Disconcordant was defined as a difference between the rhinorrhea sample and the nasal sample.
| Number (%) of Positive Samples | p Valuea | Sensitivity [95% CI] | Specificity [95% CI] | Concordanceb [95% CI] | |
|---|---|---|---|---|---|
| Nasal samplec | 79 (90%) | Reference | NA | NA | NA |
| Rhinorrhea sample | 79 (90%) | 1.000 | 97% [91%−100%] | 78% [45%−94%] | 95% [89%−99%] |
- Abbreviations: CI, confidence interval; NA, not applicable.
- a McNemar's test.
- b Concordance: proportion of true positives and negatives if true positive means both the rhinorrhea sample and nasal sample are positive for the detection of at least one pathogen and true negative means both the rhinorrhea sample and nasal sample are negative for pathogen detection.
- c The reference sample is the nasal sample, which was a (combined) mid-turbinate nasal (and oropharyngeal) sample.
3.3 Secondary outcome measures
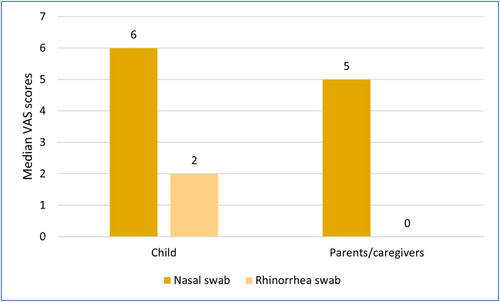
The experience of children, as reported by their parents, during the collection of an RS from nasal discharge or blowing their nose into a handkerchief, was significantly better for the RS (median VAS [IQR]: 2.00 [0.00−3.00]) than their experience with the NS (median VAS [IQR]: 6.00 [4.00−8.00]) (Wilcoxon test p = <0.001). Parents also endured the sampling of RS significantly better (median VAS [IQR]: 0.00 [0.00−2.00]) when compared with that of the NS sample (median VAS [IQR]: 5.00 [2.00−7.75]) (Wilcoxon test p = <0.001). The VAS scores are presented in Figure 2. Significant differences in total median VAS scores were found between the two groups (centers) for both the children (Mann−Whitney U test, p < 0.001) and their parents/caregivers (Mann−Whitney U test, p = 0.001). Sub analysis of VAS scores to correct for sampling order was also performed and can be found in the Supporting Information File: Table 1.
4 DISCUSSION
This study explored the performance and tolerability of two different swab methods for detecting viral pathogens in young children using multiplex PCR and SARS-CoV-2 RT-PCR. The high sensitivity of 97% (95% CI 91%−100%) and specificity of 78% (95% CI 45%−94%) demonstrates that respiratory viruses can reliably be detected by this new sampling method. In addition, the median differences in VAS score, 4.00 and 5.00 for children and their parents or caregivers respectively, demonstrate that rhinorrhea samples are better tolerated when compared to the standard sampling procedure. Previous research revealed the same conclusion, less invasive sampling procedures are better tolerated.24, 25 The overall detection rate of respiratory viruses was 92%, which was in line with another previously published study by Ippolito et al.26 Moreover, specificity is determined by false positive results, referring to pathogens that would be detected by the RS and missed by the reference standard, the NS. Considering the results of the RS as false positive would actually result in the prohibition of the RS's ability to outperform the NS with respect to pathogen detection. The use of a combined reference standard may be a solution and is described in more detail in the Supporting Information File: Table 2.6, 27
Interestingly, the RS showed an overall higher viral pathogen detection rate compared to the NS (92% vs. 89%). The RS detected 14 viral pathogens additionally to those detected by the NS, yet missed 10 viral pathogens that were found by the NS. In total there were 88 paired samples, and of these 69 (78%) completely matched each other. Disconcordance between paired samples and detection of a pathogen only by the less invasive specimen was also demonstrated in previously published literature.6, 27 Differences between paired samples could be related to quality of specimen collection, biological differences of the specimens, differences in transport medium, variability of analysis or any combination of these. The RSs were better tolerated and probably resulted in higher quality samples than the NSs. In addition, various studies indicate biological differences between different respiratory specimens. A review by Man et al. discusses that the upper respiratory tract consists of distinct micro niches colonized by microbial communities, implying viruses can reside in different micro niches.3 Furthermore, a study by Goggin et al. demonstrated that discord exists in the viral species found in the different sites of the nasal cavity.28 These studies therefore testify to biological differences between sites of sampling. A clinical study comparing NPS and OPS for the diagnosis of respiratory viruses also found that different viruses are more likely to be detected by one of both sampling methods, but also demonstrated that the less sensitive technique still detected a considerable number of viruses not detected by the higher sensitive technique.29 Perhaps, unless sampling the preferred site, differences between specimens remain and detection of respiratory pathogens can be missed. However, nasal discharge moves across different anatomical sites, probably reducing the chance of missing the detection of a certain pathogen. In addition, different swabs and transport media were used for collection of the RS and NS, which might have contributed to differences in the detection rate of respiratory viruses. Variation in analysis of both samples cannot be excluded, but is considered unlikely in this study. Nevertheless, the time between both sampling collections might also have been of influence on the differences found between both sampling methods. This can result in a possible change in viral load, due to for example individual viral load kinetics, which might have also influenced the result.30 To account for a possible influence of time between the samples, a sub analysis of the paired samples which were obtained within an hour and within 6 h was performed and is presented respectively in the Supporting Information File: Table 3 and Table 4. Sub analysis revealed comparable sensitivity, while the concordance rate for a complete match was higher, respectively 96% for 1 hour and 95% for 6 h, compared to an overall concordance of 78%.
4.1 Strengths and limitations
The present study's crucial strength, as mentioned earlier, is that the use of rhinorrhea samples taken directly from nasal discharge has not been previously studied. Therefore, the evaluation of rhinorrhea samples as a specimen for the detection of respiratory pathogens is a valuable addition to the existing literature and advances the options in this emerging field of noninvasive sampling. Furthermore, a method of sample collection was examined which is less invasive and easier to perform than a routine sampling procedure, which can aid in its capacity for implementation. Moreover, it was hypothesized that the collection of rhinorrhea samples would be better tolerated by young children and their parents, which was also objectified by the VAS score analyses. Finally, the timing of this study, which included sampling during all seasons, perhaps provided a high overall detection rate (92%) of respiratory viruses, including clinically relevant respiratory viruses, such as RSV and influenza. Another virus that became important during the past couple of years has been SARS-CoV-2 and was found in less than 7% (6/88) of the population, all diagnosed at the GGD location. However, the concordance for both sampling methods in detecting SARS-CoV-2 was 100%, meaning RSs could aid in a future pandemic when less invasive methods are required for diagnostics in children.
Despite some encouraging results, it is important to note that the sample size of 88 participants did not allow for pathogen specific conclusions. While cautious conclusions can be drawn concerning the most frequently detected viruses, such as rhinovirus and respiratory syncytial virus, no solid conclusions should be drawn yet regarding other viruses due to the small number of cases found in this population.
Additionally, different sampling methods were used during routine care and in about 1/3 of the cases it was unknown how the nasal swab was performed. The RS was therefore not compared to the gold standard, the combined NP- and OP swab, which might have led to a higher sensitivity for the rhinorrhea samples. Nevertheless, the comparison of rhinorrhea samples with these different sampling techniques is in line with clinical practice and different recommendations.16, 18-20 Also, the results should perhaps not yet be generalized to a larger population. This was a two center study and only children admitted to the emergency department, pediatric ward or tested at the Public Health Service testing facility were included in this study. Moreover, while children aged from 0 to 5 years were eligible for participation, median age of the participants was 1 year and therefore these results particularly concern a young population. Practical challenges for implementation in routine care also remain. Children presenting to the emergency department with covid-19 relating complaints, who were admitted to the pediatric ward, were tested for the presence of SARS-CoV-2 using rapid point-of-care testing. These samples were in return then used for additional diagnostics for other respiratory viruses using multiplex PCR. However, the use of rhinorrhea samples for rapid point-of-care testing has not been explored and therefore should be studied as its ability to detect viruses is absolutely present. Nonetheless, RSs might not replace nasal swabs altogether, since nasal discharge is not always present at the moment sample collection and pathogen detection is warranted. A possible solution for this, as mentioned earlier, would be to use another noninvasive sampling method such as saliva which is also considered a strong reservoir for viruses. A possible future study should therefore evaluate the performance of rhinorrhea samples in comparison to saliva samples. In addition to this, multicenter studies in the Netherlands, as well as international studies could be performed to expand the sample size and assist in detection of viral pathogens in third world countries. Moreover, the use of self-testing of nasal discharge (antigen testing) might be a possibility worth exploring, as it might lead to a higher willingness to test in both children and their parents. Lastly, the detection of viral pathogens in nasal discharge could be studied among older children or even adults to see if this could aid in diagnostics in those patient groups.
5 CONCLUSION
In summary, this study revealed that swabs taken from nasal discharge reliably detect viral pathogens in young children in both a hospital and outpatient setting. Furthermore, this alternative testing method was better tolerated by both children and their parents. In cases such as the past pandemic, it is important to note that the minor difference in sensitivity of this alternative testing method might be compensated by an increase in willingness to test. Further research is highly warranted to confirm these results and enable the implementation of rhinorrhea samples as a specimen for the diagnosis of respiratory viruses in young children.
AUTHOR CONTRIBUTIONS
Marlies A. van Houten envisioned the idea for this study. Marlies A. van Houten and Elandri Fourie wrote the study protocol and submitted a successful funding application. Marlies A. van Houten, Sjoerd M. Euser, Paul Badoux, Mildred E. Haverkort, Yara E. E. Sijm, and Elandri Fourie decided upon the methodology and execution of the sample collection. Marieke E. Mérelle, Mitchel Wit., Yara E. E. Sijm and Elandri Fourie actively collected and managed the data. Sjoerd M. Euser and Paul Badoux decided upon the execution of the laboratory analysis. Elandri Fourie and Yara E. E. Sijm performed statistical analysis and interpretation of the data and were supervised by Sjoerd M. Euser. Yara E. E. Sijm and Elandri Fourie drafted the manuscript which was then edited by all coauthors. All coauthors reviewed the manuscript and all authors have read and agreed to the published version of the manuscript.
ACKNOWLEDGMENTS
We admiringly recognize the contribution of both parents and their children in this study. In addition, we thank Dr. Marieke E Mérelle (pediatrician at Spaarne Gasthuis), Drs. Yara Sijm (YS), Mitchel Wit (MW, student) and all the pediatric ward and public health testing facility staff for their contribution in the data collection of this study.
Funding for this project was provided in the form of a grant by the scientific department of the Spaarne Gasthuis hospital (The Netherlands).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
Written informed consent was obtained from all parents or caregivers and relevant data during this period were collected retrospectively from the children's patient records. This study was assessed and ethically approved by the Medical Ethics Committee of the Vrije Universiteit Medical Center on August 4th, 2021 (VUmc; reference: 2021.0403).
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




