Single-cell transcriptomic analysis of the role of HPV16-positive macrophages in cervical cancer prognosis
Abstract
Almost all cases of cervical cancer (CC) can be attributed to high-risk human papillomavirus (HPVs) infections in keratinocytes. However, it is unknown whether HPV invades immune cells such as macrophages and T cells. We analyzed the single-cell transcriptome of the CC and its adjacent tissues and found that HPV16 genes, including E1, E6, and E7, expressed in the macrophages and CD8+ T cells in addition to the malignant cells. HPV16+ macrophages highly expressed the genes that promote cell adhesion and the favorable genes such as WAS, IQCB1, MYO1F, and PDZD11 in CC prognosis. The transcription factor KLF5 potentially accounted for the induction of these protective genes and thus facilitated the infiltration of the immune cells in tumor tissues. Our single-cell transcriptome analysis suggests the potential value of the HPV16+ macrophage in CC prognosis. However, extensive experimental studies investigating the characteristics and functions of the HPV+ immune cells are still required.
1 INTRODUCTION
Cervical cancer (CC) is one of the leading causes of death among women globally, and high-risk subtypes of human papillomavirus (hrHPV) infections have been established as the primary etiological factor of the disease in most cases.1, 2 Despite continuous efforts to improve the prevention, screening, diagnosis, and treatment of CC, the survival rate for CC patients has not been significantly improved. A deep understanding of the etiology of the disease may be critical for developing effective prevention and therapy strategies.
Li et al.3 provided the first single-cell RNA-seq atlas of CC tissue and adjacent tissue, which revealed heterogeneity of malignant cells and the development of endothelial cells across CC progress. Moreover, they designed a more detailed atlas of CC, including cervical premalignant lesion, CC, and metastatic lymph nodes, to distinguish the divergence of the immunological microenvironment, which indicated that high-stage intraepithelial neoplasia exhibited a lowly activated tumor microenvironment (TME), tumor displayed immunosuppressive state, and metastatic lymph node showed early activated phase of immune response.4 However, in these two analyses, Li et al. focused on the composition of the microenvironment and the transition between different CC stages while ignoring the influence of HPV-infected host cells in cancer.
Previous studies implied that the mortality of the hrHPV+ group was significantly lower than that of the hrHPV− group, indicating that hrHPV infection might confer a better prognosis in CC.5, 6 A similar phenomenon was observed in head and neck squamous cell carcinoma (HNSCC), especially in oropharyngeal cancer.7, 8 The infiltration of immune cell populations appeared to have different patterns in HPV+ HNSCC and HPV− HNSCC, including a higher frequency of intra-tumoral B cells, IFNγ+ CD8+ T lymphocytes, and IL-17+ CD8+ T lymphocytes in HPV+ HNSCC,9-11 and a higher frequency of dysfunctional CD8+ T cells in HPV− HNSCC.12 It is acknowledged that integrating the HPV genome into the host cells induced immune evasion of host cells.13 Meng et al.14 reanalyzed the CC's data sets and proposed some evidence to explain the reasons why the detectable hrHPV lead to a better prognosis. Detailly, HPV+ malignant cells showed decreased stemness, while HPV− malignant cells had detrimental interactions with the endothelial cells.14 Although they had listed some evidence that HPVs might decrease the expression of a set of unfavorable molecules in the HPV+ malignant cells, including KRT16, ITGB1, CXCR1, VEGFA, CRCT1, and TNFRSF10B/DR5, whether HPVs have other effects on immune cells remains unclear.
Interestingly, Li et al.3 and Meng et al.14 both demonstrated that there were some immune cells to be infected by HPV by detecting the expression of TP53/RB1 and HPV transcripts, respectively. HPV+ immune cell populations are special immunological features in HPV+ tumors. To date, the potential functions of the HPV+ immune populations in the development and progression of CC are not clear, which needs to be further investigated. Based on the above findings, we collected HPV+ cells from CC and tried to determine whether HPV+ immune cells are favorable factors in hrHPV-associated cancers.
2 MATERIALS AND METHODS
2.1 Collection and processing of the CESC RNA-seq data sets
The RNA sequencing data set and the clinically related data for patients with CSCC originated from the TCGA database (GDC TCGA Cervical Cancer [CESC], http://xena.ucsc.edu/) and consisted of 296 samples. The FPKM gene expression data set was downloaded from the website. Samples with missing data were excluded. The scRNA-seq data from patients with CC originated from the gene expression omnibus database and were accessed through NCBI GSE168652.3 Raw fastq data of scRNA-seq were processed using UMI-tools,15 and viruses present in the single cells were detected using the Viral-Track approach.16 In brief, the sequencing data containing the single cell index were mapped to the virus genome reference database and the status of the single cell was added to the expression matrix to correlate with the presence of HPV infection and the corresponding transcriptome.
2.2 Single cell RNA sequencing analysis
The scRNA-seq data sets were processed using Seurat (version 4.1.1).17 In detail, the filtration aimed to collect high-quality cells (the number of genes 200−6000; the percentages of mitochondrial genes <25%). The gene expression was normalized. The top 2000 variable genes were scaled and used for principal components analysis (PCA) dimensionality reduction. After that, two data sets were integrated. PCA analysis was further conducted for the integrated data sets, and cluster analysis was performed by using uniform manifold approximation and projection and t-distributed stochastic neighbor embedding (t-SNE).
2.3 Differentially expressed genes (DEGs) analysis
DEGs were calculated using FindMarkers in Seurat. In detail, genes should express at least 25% of cells, and the log-transformed fold change should be higher than 0.25. To calculate specific features in HPV+ immune cells, the ratio difference of genes in two clusters should be up to 0.5. Moreover, the ratio of each gene in all cell types was calculated. Features with ratios up to 0.35 in any cell type were removed.
2.4 Function enrichment analysis
The gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed using Metascape with default parameters.18 Only GO terms and KEGG pathway data sets were referenced.
2.5 Kaplan−Meier (KM) survival analysis
KM survival analysis was performed using the R packages survival19 and survminer.20 The genes with higher expression than mean expression were in the highly expressed group, and others were in the low expressed group. Using the Gρ family of tests to test if there is a difference between two survival curves.
2.6 Cell to cell communication
The CellPhoneDb v4 was used for constructing communication between different cell types using default parameters.21 All ligand-receptor pairs with p value less than 0.05 were retained.
2.7 Gene regulatory network construction
The gene regulatory network was constructed using pySCENIC.22 In detail, we used the singularity to run the image of pySCENIC (version 0.12.0). First, we used “GRNBoost2” to obtain the TFs and their target genes, defining the regulons. The regulons were generated using gene inference methods, which solely rely on gene expression correlations across cells. Second, the regulons were refound by pruning targets, which were absent in the enrichment of a corresponding motif. TF effectively separated from d direct or indirect targets based on cis-regulatory footprints. Finally, the original cells were assigned and clustered according to the activity of these discovered regulons. The regulon activities were binarized based on the bimodal model. After getting the matrix of different regulon activities, we calculated the proportion of activated cells in each subtype to create a new activity matrix. The UpSet23 was used for visualizing the intersection of target genes between different regulons.
3 RESULTS
3.1 Detection of HPV16 infection in CC
CC data sets were collected from a 58-years HPV16+ patient, including the tumor tissue and its adjacent normal tissue. During the data mining, we clustered all cells as 11 clusters which were annotated as seven main cell types (Figure 1A; Supporting Information: Figure S1). Malignant cells (clusters 0, 2, 3, 8) highly expressed epithelial markers, including CDH1, EPCAM, and CDKN2A. KRT14, KRT15, and KRT16 were also highly expressed in malignant cells. Endothelial cells (cluster 5) highly expressed PECAM1, EMCN, and CD34. CD8+ T cells (cluster 9) showed specific expression of CD3D, CD3E, CD3G, CD8A, and NKG7. Macrophages (cluster 10) were identified based on CD163, CD14, and MRC1. Cancer-associated fibroblast (CAF, cluster 6) expressed FAP and COL1A2 specifically. Other clusters all displayed the characteristics of smooth muscle cells, including ACTA2, CNN1, MYH11, and DCN, while cluster 4 and cluster 7 also expressed PDGFRB and MCAM specifically. Therefore, cluster 1 was annotated as smooth muscle cells, while cluster 4 and cluster 7 were annotated as vascular smooth muscle cells (Figure 1B).

We used UMI-tools15 and Viral-Track16 to detect the expression of HPV16 genes. Analysis results displayed that HPV16 was mainly expressed in malignant cells, CD8+ T cells, and macrophages (Figure 1C). The reads mapped to the HPV16 genome were enriched in the E1, E6, and E7, which played critical roles in transcriptional dysregulation of the infected host cells.24 Devitt et al.25 emphasized some existing problems in the alignment from viral transcripts to virus genome due to the technology, including more mapped reads in E5 but not in other regions. However, no reads were mapped in E5 in our data sets.
HPV is also one of the causes of HNSCC. Cillo et al.26 analyzed the divergency of the immune microenvironment between HPV+ HNSCC and HPV− HNSCC. We reanalyzed their data set and found that one data set detected the existence of HPV. In this data set, we confirmed that there were some HPV+ macrophages and T cells in this data set (Supporting Information: Figure S2). We got 3790 cells from the data set (SRR10340992) (Supporting Information: Figure S2A). The cluster 6 was identified as macrophages that highly expressed CD86, CD14, and LYZ. Most of the other clusters were identified as T cells which highly expressed CD3 (Supporting Information: Figure S2B). Then, there were some cells that expressed HPV16, including macrophages and T cells (Supporting Information: Figure S2C−D).
3.2 Increased cell adhesion ability of the HPV16+ macrophages
Next, we aimed to further characterize the HPV16+ immune cells and investigate their potential functional changes. Tumor-associated macrophage (TAM) has high plasticity and plays an important role in regulating immune function in TME.27 The infiltration of TAM is correlated with the CC progress.28 The macrophages were isolated from the merged data set and were divided into three groups based on the expression HPV16 (Figure 2A). To ensure the reliability of cell annotation, we plotted the typical macrophage markers in the HPV16+ macrophages (Figure 2B). Almost all cells expressed at least one of the markers. Considering the drop-out in scRNA-seq, we determined these cells as the HPV16+ macrophages.
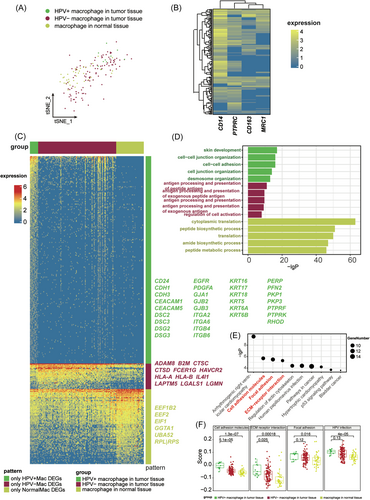
We calculated the significantly DEGs in these three groups and performed GO term enrichment analysis (Figure 2C,D). HPV16+ macrophages displayed increased activity in intracellular adhesion. Detailly, HPV16+ macrophages specifically expressed a huge amount of cell adhesion molecules, including CD24, Cadherin (CDH1/3, CEACAM1/5, DSC2/3, DSG2/3), gap junction protein (GJA1, GJB2/3), integrin (ITGA2/6, ITGB4/6), keratin (KRT5/6A/6B/16/17/18), plakophilin (PKP1/3), protein tyrosine phosphatase receptor (PTPRF/K), RHOD, PERP, PEGFRA, and EGFR (Figure 2C). Specific DEGs in the HPV16+ macrophages were significantly enriched in adhesion-associated terms (Figure 2D). The phenomenon suggested that HPV16+ macrophages might have more interaction with other populations in tumor tissue compared to the HPV16− macrophages.
KEGG pathway enrichment also indicated that cell adhesion molecules were significantly upregulated in the HPV16+ macrophages (Figure 2E). To avoid the misjudgment that the upregulation of cell adhesion was due to increased expression of a few specific genes, we collected all genes belonging to the pathway “ECM−receptor interaction,” “Focal adhesion,” and “Cell adhesion molecules,” and estimated each pathway score based on the expression of all genes in each pathway. Specifically, we binned features based on average expression and randomly selected 100 control features from each bin. Aggregated expression of gene set features was subtracted by the aggregated expression of the control feature set to obtain a gene set score. KEGG pathway scores demonstrated that HPV16 infection promoted cell adhesion expression (Figure 2F). In conclusion, HPV16 infection appears to increase macrophage adhesion, which might influence the infiltration of macrophages. We also checked the divergence between HPV16+ CD8+ T cell and other cells. HPV16+ CD8+ T cell also displayed increased cell adhesion (Supporting Information: Figure S3).
3.3 HPV16+ macrophages highly expressed favorable factors during CC prognosis
Even though HPV16+ macrophages showed functional divergence in cell adhesion compared to HPV16− macrophages, the role of HPV16+ macrophages in CC progress is unclear. To conduct whether the HPV16+ macrophage is a favorable factor in CC prognosis, we calculated all significant DEGs of HPV+ macrophages compared to other cells. We summarized a set of genes that are specifically expressed in HPV16+ macrophages from DEGs (Figure 3A). To investigate which gene is the favorable or unfavorable factor to CC prognosis, we did the KM survival analysis using CESC data sets from TCGA. Finally, there were four genes that were significantly involved in the prognosis, and all of them are favorable factors for a better prognosis, including WAS, IQCB1, MYO1F, and PDZD11 (Figure 3B). Taken together, our results indicated that HPV16+ macrophages might be a favorable factor for a better prognosis.
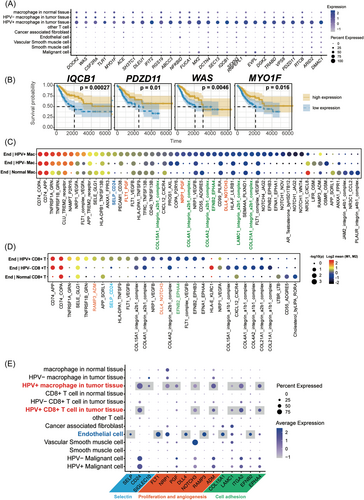
To check whether HPV16+ CD8+ T cell is also a favorable factor for CC, we tried to find out the specifically expressed genes in HPV16+ CD8+ T cells. However, no gene was specific enough. To further verify whether the four favorable features are also different in HPV+ macrophages than macrophages in HPV− CC tumors, we found a data set (GSE171894) that contained more than 30 000 cells from HPV16+ and HPV16− CC patients. We selected two HPV16− data sets and determined cluster 6 as macrophages (Supporting Information: Figure S4A−B). We checked the four favorable features and found that IQCB1 and PDZD11 are significantly higher in the HPV16+ macrophages than the macrophages in the HPV16− tumor. The result further confirmed the divergence between HPV16+ macrophages and HPV16− macrophages.
T cells homing from the bloodstream into normal tissues or tumors require three distinct steps: selectin-mediated T cell rolling on endothelium, chemokine-mediated activation of integrins, and T cell transmigration dependent on activated integrins.29 Then, we used CellPhoneDB to investigate the communication between endothelial cells and HPV16+ macrophages/CD8+ T cells.30 Compared to the interaction between HPV16− macrophages/CD8+ T cells and endothelial cells, HPV16+ macrophages/CD8+ T cells had more specific interactions with endothelial cells (Figure 3C,D). For example, the SELP-CD24 pair was significantly activated in HPV16+ macrophages/CD8+ T cells to endothelial cells (Figure 3C,D). As shown in Figure 3E, SELP, the selectin P, is highly expressed in endothelial cells, representing the signal of recruitment to immune cells. HPV+ immune cells, including macrophages and CD8+ T cells, both highly expressed CD24, indicating that HPV16+ immune cells might be easier recruited to the tumor region through the blood vessels. Besides, malignant cells also dramatically expressed CD24 and SIGLEC10 (Siglec-10), specifically expressed in macrophages, indicating that CD24-Siglec-10 interaction blockade might be a promising therapeutic strategy for CC immunotherapy (Figure 3E).
FLT1-PGF and NRP1-PGF were shown to be specifically activated in HPV16+ macrophages to endothelial cells. NOTCH3 is expressed in malignant cells, vascular smooth muscle cells, and HPV16+ immune cells broadly (Figure 3D), suggesting stronger proliferation in these cells. Moreover, Ramp3_ADM, which was involved in the angiogenesis,31 specifically activated between endothelial and HPV16+ immune cells (Figure 3C,D). Another specific set of interactions is associated with cell adhesion, including the interaction between collagens/Laminin and Integrin alpha-2/beta-1 (Figure 3C). The expression of ITGA2 was more abundant in malignant and HPV16+ immune cells (Figure 3D). EphrinB2/EphA4-mediated activation of endothelial cells could increase the monocyte adhesion.32 HPV16+ immune cells showed stronger expression of EPHA4, indicating that HPV16+ immune cells might play a pivotal role in the development of recurrent, invasive, and distant metastasis. In conclusion, our results suggested that HPV16 infection might increase the recruitment of immune cells to endothelial cells and increase cell adhesion.
3.4 HPV16 might regulate the infiltration of immune cells mainly through KLF5
To investigate the potential mechanism of how HPV influences host immune cells, we used pySCENIC to construct the regulatory relationship between transcription factor (TF) to targets.22 Detailly, pySCENIC aims to infer TF-targets sets (regulons) with relatively higher activity in one cell type compared to other cell types, which is mainly based on coexpression association between TFs, targets, and TF binding motif in the promoter region of the targets. Based on the complex gene regulatory network, we hoped to determine whether there were TFs involved in the transcriptional regulation of HPVs infection. Finally, we obtained 374 useful regulons with different expression patterns in different cell types. The regulons which contained the four favorable genes in CC prognosis were selected (Figure 4A). WAS and IQCB1 were involved in more than 10 regulons (Figure 4A). The phenomenon that a gene is involved in many regulons suggested that the gene could be regulated by many TFs. Since genes in a regulon might be functionally related to each other, participating in more regulons suggests that the gene has a more complex function. WAS and IQCB1 were involved in the most regulons. There was a set of regulons that displayed stronger activity in HPV16+ immune cells than other immune cells, including BCL11A, TFDP1, MBD2, KLF5, EHF, and PITX1. Among these TFs, the expression of BCL11A, TFDP1, KLF5, EHF, and PITX1 were consistent with the regulons' activity, suggesting that these TFs might be influenced by HPV16 infection (Figure 4B). WAS and MYO1F belonged to the regulons of IKZF1, ZNF331, SPI1, IRF8, ETV5, MAFB, and CREM. However, these TFs seemed to negatively regulate the expression of WAS and MYO1F. The adjacencies between TFs and targets reflected that KLF5 and EHF had the highest adjacencies with IQCB1, while SPI1 and IKZF1 had the highest adjacencies with WAS and MYO1F (Figure 4C). The phenomenon suggested that the four TFs might be the critical factor that regulated IQCB1, WAS, and MYO1F in HPV16+ macrophages.
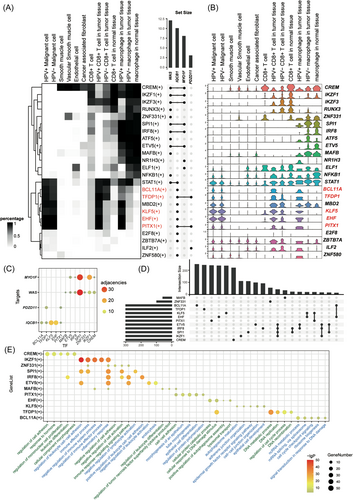
To investigate the biological function of regulons, target genes with the highest adjacencies in each TF were selected for functional enrichment analysis. Gene sets intersection displayed that most targets of regulon are unique in each regulon, suggested that each regulon might had its unique characteristic and function (Figure 4D). GO term enrichment displayed that CREM(+) was associated with cell adhesion and cell proliferation. IKZF1(+), SPI1(+), IRF8(+), MAFB(+), and ETV5(+) mainly participated in similar biological progress, including leukocyte adhesion, inflammatory response, and negative regulation of cell activation. PITX1(+) was involved in the morphogenesis of the epithelial tube. KLF5(+) was related to the epidermal growth factor receptor signaling pathway and adhesion-associated function. BCL11A(+) was involved in RNA processing and cell cycling, while EHF(+) was associated with DNA damage response (Figure 4E). Hu et al.33 provided a detailed reference describing the HPV16 integration points in the human genome. Both sanger verification of DNA and RNA confirmed that there were several breakpoints between KLF5 and KLF12, suggesting the affection of HPV16 integration into the genome in host cells. These data together indicated that partial specific transcriptional changes in immune cells due to HPV16 infection might arise from the change of KLF5.
4 DISCUSSION
Persistent infection with hrHPV has emerged as a major and necessary etiological factor for CC and other HPV− related malignancies. Despite continuous efforts that have been made, the entire mechanisms of disease progression remain unclear. On the one hand, it is assumed that HPV infection can modulate the microenvironment and evade the host immune responses.1 On the other hand, several studies have indicated that the presence of HPV can lead to a more favorable prognosis.14, 34 Therefore, the aim of the current study was to determine the potential role of the HPV+ cells, particularly the HPV+ immune cells, in the CC prognosis by taking advantage of the single-cell transcriptomic analysis.
First, we used Viral-Track16 to estimate HPV16 infection in CC. Interestingly, we found that in addition to being abundantly expressed in malignant cells, HPV16 can also be expressed in macrophages and CD8+ T cells. Previous studies have shown that HPV primarily infects poorly differentiated basal keratinocytes,35 raising the question of whether these HPV+ immune cells might exert distinct roles to that of their HPV- counterpart in the progression of CC. Therefore, we want to further characterize the HPV+ immune cells and investigate their underlying functional changes.
Macrophages were isolated from the merged data set and divided into three groups based on the expression levels of HPV16 oncoproteins (E1, E6, and E7). Significant DEGs in these above groups were calculated, and functional enrichment analyses were then performed to investigate the potential functions of HPV16+ macrophages. It is worth noting that specific DEGs in HPV16+ macrophages significantly enriched in adhesion-associated terms. Consistent with the GO analysis results, KEGG pathway analysis showed that DEGs in HPV16+ macrophages were highly enriched in “cell-cell adhesion,” indicating that HPV16 infection might enhance macrophages adhesion, modulate the physiological interactions between HPV16+ macrophages and other populations in the TME. Besides, increased adhesion might assist HPV16+ macrophages to cross barriers and promote the tissue infiltration of the macrophages.36, 37
Next, KM survival analysis for the above significant DEGs was further conducted to explore the potential effects of HPV16+ macrophages in CC progression. Among the 27 candidates, 4 genes (WAS, IQCB1, MYO1F, and PDZD11) were shown to be significantly positively related to the overall survival of CC patients. A lot of research reported that the IQCB1 and PDZD11 expressions positively correlate with macrophages and CD8+ T cell infiltration in colon cancer.20, 21 WAS is an important regulator of the actin cytoskeleton in hematopoietic cells and lymphocyte homeostasis.22 MYO1F was involved in the M1-polarization of macrophages.23 Taken together, these results indicated that HPV+ macrophages might be a favorable factor for a better prognosis.
CellPhoneDB analysis was then performed to infer the communication between the endothelial cell and HPV16+ macrophages/CD8+ T cells from a combined expression of ligand-receptor pairs. Our findings revealed that compared to the interaction between HPV16− macrophages/CD8+ T cells and endothelial cells, HPV16+ macrophages/CD8+ T cells had more specific interactions with endothelial cells. For example, CD24, which has been reported to act as a ligand for P-selectin, was shown to been highly expressed in HPV+ macrophages and CD8+ T cells. Activation of the SELP-CD24 pair might promote the recruitment of HPV16+ immune cells to the tumor region through the blood vessels. We also observed a high expression level of CD24 in CC cells, which has been identified as a dominant innate immune checkpoint in several cancers through interaction with macrophage Siglec-10,38 suggesting that therapeutic potential for CD24-Siglec-10 interaction blockade in CC immunotherapy. Another specific set of interactions between HPV16+ macrophages/CD8+ T cells and endothelial cells was identified to be associated with cell adhesion. Previous studies have demonstrated that the adhesion of immune cells to endothelium is vital for immune surveillance.39, 40 Considered together, HPV16 infection could increase the recruitment of immune cells to endothelial cells and promote cell adhesion, which may assist HPV16+ immune cells in trafficking across the vascular endothelium. It may be suggested that HPV16 infection might promote the switch from an immunologically “cold” TME to a better infiltrated “hot” TME, with enhanced immune cells infiltration in the tumor core.41
Intriguingly, specific activation of FLT1-PGF and NRP1-PGF pairs was found between HPV16+ macrophages and endothelial cells. PGF secreted by HPV16+ macrophages might signal to endothelial cells through surface expressions of FLT1 and NRP1 on endothelial cells, which may contribute to endothelial cell proliferation and angiogenesis.42, 43 Similarly, Ramp3-ADM, which was involved in the angiogenesis,31 has also been shown to be specifically activated between endothelial and HPV16+ immune cells. Angiogenesis is a key factor that favored the persistence, integration, and progression of cervical neoplasia.24 However, T cells homing from the bloodstream into tumors also depend on selectin in vascular endothelial cells.29 Angiogenesis may provide a positive effect on the HPV16+ immune cell infiltration while providing nutrients for malignant tumor cells.
Genetic regulatory programs control the immune responsiveness.44 The ability to elicit gene-expression changes in macrophages is influenced by the accessibility of TFs binding sites in the macrophage genome.45 However, the characterizations of gene regulatory programs of HPV16+ macrophages in CC are still unclear. We further extended our studies to systematically investigate the potential underlying mechanisms for the influences of HPV16 infection on host immune cells. pySCENIC analysis was performed to construct the gene regulatory network. Detailly, pySCENIC aims to infer TF-target sets (regulons) with relatively higher activity in one cell type compared to other cell types, which is mainly based on coexpression association between TFs and targets and TF binding motif in targets' promoter region. Our current data found 12 important TFs which might have an influence on the function of macrophages and suggested that HPV16 might increase the adhesion of immune cells mainly through KLF5 and EHF. KLF5 expressed in malignant cells could promote CC proliferation, migration, and invasion.46 Besides, the inhibition of EHF might impair the cell adhesion.47 Our findings further indicated the different roles of KLF5 in immune cells, which might be positive in immune infiltration.
The process of HPVs infecting host cells is related to interaction between capsid protein and heparan sulfate proteoglycans (HSPG) in base membrane (BM).48 L1 contains the major determinants for HPV attaching to host cells. In the BM, HPV binds to HSPG, which induces a conformational change exposing a site on L2 susceptible to proprotein convertase cleavage. Then, an exposed L2 neutralizing epitope and a previously unexposed region of L1 bind to a secondary receptor on the invading edge of the epithelial cells, which induces the HPV infection in vivo. Binding to the BM may promote preferential interaction with basal keratinocytes, which leads to keratinocytes being the main targets of HPV. Although several types of immunocytes bind and internalize PV capsids, including monocytes and macrophages, there is no evidence that the interaction results in infection of these cell types either in vitro or in vivo.
Local signals enforce distinct functional states of macrophages, of which two ends of a spectrum of activation (M1 to M2) are commonly recognized.49 M1 macrophages are involved in efficient antigen presentation and pathogen killing. M2 macrophages show high phagocytic activity. They contribute to inflammation resolution. Whether the emergence of HPV transcripts is through unknown receptors or the phagocytic activity of macrophages requires more research.
In conclusion, this study illustrated that HPV+ immune cells could exert strikingly distinct roles from that of their HPV− counterparts in the progression of CC. HPV infection might enhance immune cell adhesion and promote the infiltration of these HPV+ cells into the tumor core region. Further work will be required to unravel the precise roles of HPV+ immune cells in the development and progression of CC.
AUTHOR CONTRIBUTIONS
Feng Ma conceived the idea. Shiyou Wang performed data analysis and wrote the manuscript draft. Xiaohui Li and Chao Liu provide helpful discussions and key resources. Feng Ma and Yuan Yi revised and edited the manuscript, and all authors approved the final version of the paper.
ACKNOWLEDGMENTS
This study was supported by the National Key Research and Development Program of China (2018YFA0900803), National Natural Science Foundation of China (32170880), Natural Science Foundation of Jiangsu Province (BK20200004 and BK20210118), CAMS Innovation Fund for Medical Sciences (CIFMS2021-I2M-1-047 and CIFMS2022-I2M-2-004), and Non-profit Central Research Institute Fund of CAMS (2019PT310028).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
All data sets used in this research could be found in the GEO database (HPV+ CC: GSE168652; HPV− CC: GSE171894; HPV+ HNSCC [GSE139324-SRR10340992]) and TCGA database. Further inquiries can be directed to the corresponding author.




