Avian influenza A virus H7N9 in China, a role reversal from reassortment receptor to the donator
Jun He, Sai Hou, Chenglong Xiong, and Linjie Hu contributed equally to this study.
Abstract
Reassortment can introduce one or more gene segments of influenza A viruses (IAVs) into another, resulting in novel subtypes. Since 2013, a new outbreak of human highly pathogenic avian influenza has emerged in the Yangtze River Delta (YRD) and South-Central regions of China. In this study, using Anhui province as an example, we discuss the possible impact of H7N9 IAVs on future influenza epidemics through a series of gene reassortment events. Sixty-one human H7N9 isolates were obtained from five outbreaks in Anhui province from 2013 to 2019. Bioinformatics analyses revealed that all of them were characterized by low pathogenicity and high human or mammalian tropism and had introduced novel avian influenza A virus (AIV) subtypes such as H7N2, H7N6, H9N9, H5N6, H6N6, and H10N6 through gene reassortment. In reassortment events, Anhui isolates may donate one or more segments of HA, NA, and the six internal protein-coding genes for the novel subtype AIVs. Our study revealed that H7N9, H9N2, and H5N1 can serve as stable and persistent gene pools for AIVs in the YRD and South-Central regions of China. Novel AIV subtypes might be generated continuously by reassortment. These AIVs may have obtained human-type receptor-binding abilities from their donors and prefer binding to them, which can cause human epidemics through accidental spillover infections. Facing the continual threat of emerging avian influenza, constant monitoring of AIVs should be conducted closely for agricultural and public health.
1 INTRODUCTION
Influenza A viruses (IAVs) can infect humans and animals causing serious global public health security concerns. Several subtypes of avian influenza A virus (AIV), such as H5N1 and H7N7, have led to high morbidity and mortality rates in poultry. In recent years, several AIV subtypes have arisen in sporadic outbreaks in humans, including H5N1, H9N2, H7N7, and so forth.1-3 These AIVs usually do not cause pandemics because of low transmissibility among humans. Unfortunately, the AIV H7N9, a novel IAV reassortant, has posed substantial risks to human health since the first emergence in the Yangtze River Delta (YRD) of China in the spring of 2013.4 Thus far, six seasonal epidemic waves have been documented in China's mainland.5 According to the statistics from the Food and Agriculture Organization of the United Nations (FAO), as of June 2, 2021, there had been 1568 laboratory-confirmed human cases of AIV H7N9 infections in China.6 Laboratory-confirmed human infections caused by H7N9 had surpassed H5N1 viruses despite its limited dissemination outside of China.7 Anhui province, located in YRD of China, is one of the regions that reported the earliest confirmed human cases of H7N9 infection.8 As of June 30, 2019, there have been five epidemic waves in Anhui province. A total number of 102 laboratory-confirmed human cases have been reported of which 52 were fatal. The number of confirmed cases in the fifth wave significantly exceeded the previous four.
A pandemic influenza virus can emerge through different mechanisms and an understanding of this will affect the scope, the focus of surveillance, and prevention efforts.9, 10 A series of reassortment events could introduce and adapt one or more genomic segments of IAVs into another one, resulting in novel subtypes of IAV, which could be a possible mechanism for generating a pandemic strain.11, 12 Three IAVs caused major pandemic outbreaks in the 20th century, namely, the 1918 H1N1 virus, the 1957 H2N2 virus, and the 1968 H3N2 virus.13, 14 The epidemics caused by H7N9 and (H1N1) pdm09 viruses are of great global concern in this new millennium.15, 16 All these viruses were generated using gene reassortment among swine, avian, and pre-existing human viruses.13-16 Until now, the H7N9 epidemic has lasted for 8 years. Although human infection is sporadic, it exists in poultry in the YRD and South-Central regions of China and maintains a high number of circulation records. In this study, we isolated 61 H7N9 strains from five epidemic waves in Anhui province from 2013 to 2019 and discussed their role and significance in generating novel subtypes of IAV through gene reassortment to support future monitoring and further prevention and control strategies for influenza epidemics.
2 MATERIALS AND METHODS
2.1 Sample collection and subtyping
The epidemiological information on avian influenza A (H7N9) cases, including date of onset and residential address, was obtained from the China Information System for Disease Control and Prevention. Respiratory specimens from suspected H7N9 patients were collected by local Centers for Disease Control and Prevention, according to the National Influenza Surveillance Guidelines. All samples were delivered in a viral transport medium with antibiotics and refrigerated before being sent to the laboratory of the National Influenza Surveillance Network. Viral RNA was extracted using the RNeasy Mini Kit (Qiagen), according to the manufacturer's instructions. The operation was performed in a biosafety level laboratory-2 (BSL-2) biohazard hood facility with personal protective equipment at the BSL-3 level. Multiplex real-time reverse transcription PCR (RT-PCR) was performed using the SensiFAST Probe Lo-ROX One-Step Kit (Bioline) on the ViiATM 7 system (Thermo Fisher Scientific). Real-time RT-PCR assays for detecting subtypes H5, H7, and H9 were performed using specific primers and probes.4, 17
2.2 Virus isolation and genome sequencing
For virus isolation, H7N9-positive samples were propagated in 9–11-day-old specific pathogen-free embryonated chicken eggs for 48–72 h at 35°C. After incubation, allantoic fluid was harvested and tested for HA agglutination. All operations were performed in a BSL-3 laboratory.
The RNAs of H7N9 viruses extracted from the allantoic fluid were selected for Sanger sequencing. Gene segments were amplified using a Qiagen OneStep RT-PCR Kit (Qiagen). Most strains were sequenced by the Chinese National Influenza Center, according to their previous study.18 For the remaining subtypes, 28 primer pairs were used to generate PCR amplicons between 176 and 923 bp in length for full genome sequencing. Amplicon purification and sequencing were conducted by the Shanghai Bio-Germ Biological Technology Co. Ltd.
2.3 Phylogenetic trees and amino acid residue signature analysis
Nucleotide sequences were assembled using Lasergene (version 7.1.0) and deposited in the Chinese influenza virus genetic sequence database and Global Initiative on Sharing All Influenza Data (GISAID). Phylogenetic tree and molecular evolutionary analyses were conducted using MEGA version 7.0.26,19 using the neighbor-joining method with bootstrap analysis (1000 replicates). Evolutionary distances were computed using the maximum composite likelihood method, with units of nucleotide substitutions per site per year. The necessary reference sequences available from the National Center for Biotechnology Information (NCBI) and GISAID were also included in the phylogenetic tree and amino acid residue signature analysis.
2.4 Reassortment events detecting and confirming
Identities of eight gene segments of 61 H7N9 isolates from Anhui province were calculated such the identities of their gene sequences retained in each segment for subsequent analysis were less than 99.5%. For each retained sequence, Basic Local Alignment Search Tool (BLAST) was performed using NCBI and GISAID. The maximum target sequences were set to 100. The sequences matrix of each BLAST was downloaded, the virus names of each sequence were extracted, and the repeated sequences were removed. The remaining names were unique. The whole genome of each virus was obtained from NCBI or GISAID. If the genome was incomplete, the virus was discarded.
Following alignment, the gene segments affiliated with the same virus were spliced individually in the order of PB2-PB1-PA-HA-NP-NA-MP-NS; for H7Ny, the splicing mode was PB2-PB1-PA-HA-NP-MP-NS; and for HxN9, the splicing mode was PB2-PB1-PA-NP-NA-MP-NS. The splicing mode for the other subtypes was PB2-PB1-PA-NP-MP-NS. All genomes were assembled into three long-sequence matrices according to their splice mode. The 61 H7N9 isolates established from Anhui province were aligned, spliced in the same manner, and merged into the corresponding long sequence matrix according to H7Ny, HxN9, and other subtypes. The alignment program was repeated for each matrix.
Reassortment events were detected using the Recombination Detection Program (RDP) Beta 4.94 (http://web.cbio.uct.ac.za/%7Edarren/rdp.html).20, 21 Seven methods of RDP, GENECONV, Bootscan, Maxchi, Chimaera, SiScan, and 3seq in the RDP package were performed with the same parameters (p < 0.01, Bonferroni corrected). Splits tree used the pairwise homology test to statistically test for the presence of conflicting signals of recombination (hereby reassortment) in a set of aligned sequences with performance similar to that of coalescence-based methods. A p < 0.05 was considered a significant signal for reassortment. When a recombination signal can be detected by no less than 5 methods, it is recorded as a possible recombination event, and further verification is carried out by using Simplot. Simplot 3.5.1 (https://sray.med.som.jhmi.edu/SCRoftware/SimPlot/)22 was used to confirm each event by using the following parameters: 700-bp window, 4-bp step size, GapStrip: on, 100 repetitions significant bootstrap >90% and Kimura (two parameters) T/t of 2.0. The genome of A/Shanghai/02/2013(H7N9) (NC_026422–NC_026429), the RefSeq for species of IAVs H7N9, was set as the outgroup, when necessary. The curves of genome identities were also plotted using Simplot 3.5.1.23, 24 The flow of reassortment event detection is summarized in Figure 1.
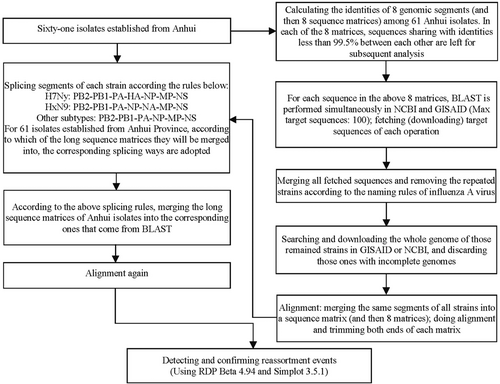
3 RESULTS
3.1 Epidemic features and IAV isolates
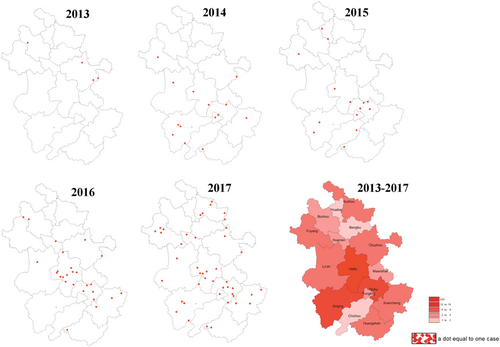
From 2013 to 2019, there were five outbreak waves of human H7N9 influenza in Anhui province with 102 laboratory-confirmed cases. They were distributed as follows: 4 cases in 2013, 14 in 2014, 14 in 2015, 26 in 2016, and 44 in 2017. No human H7N9 avian influenza cases were reported in 2018 and 2019. The cases were geographically scattered in 16 cities of Anhui province, primarily in Hefei (22, 21.57%), Wuhu (14, 13.73%), Anqing (10, 9.80%), Chuzhou (8, 7.84%), and Xuancheng (8, 7.84%) (Figure 2). Compared with the previous four waves, the fifth outbreak occurred earlier in the season, with more cases (44 in the fifth vs. 58 in the other four) and a lower fatality rate (45.90% in the fifth vs. 57.89% in the other four) (Figure 3A).
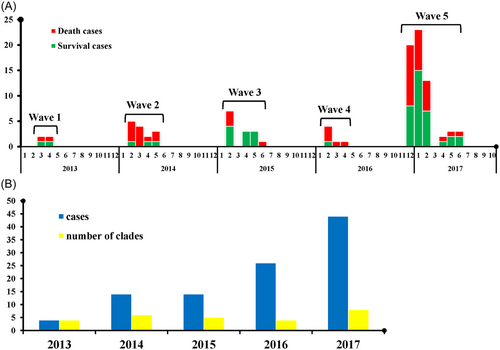
A total of 61 H7N9 isolates were established from clinical samples of 102 laboratory-confirmed human infections. Whole genomes of these isolates were deposited in the GISAID database (Table S1).
3.2 Phylogenetic features and key amino acid substitutions
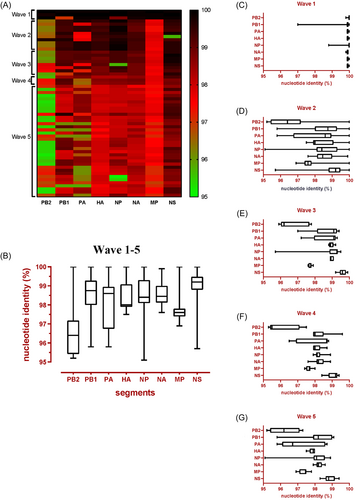
Compared with the reference A/Anhui/1/2013(H7N9) (EPI_ISL_138739), the HA and NA genes of the other 60 isolates shared more than 97% identity at the nucleotide level. However, their six internal genes exhibited distinct heterogeneity (Figure 4A); sequences PB2, PB1, PA, NP, MP, and NS of the 60 isolates shared 95.2%–100%, 95.8%–100%, 95.8%–100%, 95.1%–100%, 96.9%–100%, and 95.7%–100% identities, respectively (Figure 4B). In Wave 1, only the PB1 and NP genes were considerably different (Figure 4C). For the subsequent four waves, significant differences were also revealed in the PB2, PB1, and PA genes (Figure 4D–G). Based on the genomic diversity of the six internal genes, these H7N9 isolates could be divided into 26 clades, of which 16, 17, and 7 clades were detected in cured cases, death cases, and both, respectively (Figures 3B and S1).
For the HA gene, all of the 61 H7N9 isolates established from Anhui province were characterized by a single amino acid “R” or “K” in the HA cleavage site, indicating low pathogenicity in poultry. All the isolates possessed the G186V, T160A (150-loop), and Q226L/I (210-loop) substitutions (according to the H3 subtype), which may increase the human receptor binding of the viruses.4, 25 Other substitutions such as S138A and T160A in the HA protein associated with receptor specificity were also discovered.25, 26
The human-adaptation-associated amino acid markers in the PA protein were as follows: PA-100A was found in human isolates in every wave. K356R and S409N mutations occurred in almost all human isolates in Anhui province.27 Mutations associated with increased virulence and polymerase activity, such as M1-30D, M1-215A, M1-41A, and NS1-42S, were found in all human isolates in Anhui.28-30 Mutations associated with mammalian adaptation, such as PB1-368V, PB2-535L, and PB2-588V, occurred in most isolates, particularly in Wave 5.31, 32 PB2-E627K and D701N increased replication efficiency in cell culture and enhanced virulence in mice.33 In this study, PB2-627K was detected in most human isolates from Anhui. Meanwhile, PB2-K526R enhanced the 627K and 701N functions.33, 34 Such a substitution occurred largely in the fifth wave. No antiviral drug-resistant to neuraminidase inhibitor-related substitutions in the NA protein were detected in our isolates derived from the Anhui province. Unfortunately, they may have reduced susceptibility to rimantadine and amantadine, owing to the 31N signature in their M2 proteins (Table 1).30
| Gene | Function | Mutation | Amino acid | First wave (n) | Second wave (n) | Third wave (n) | Fouth wave (n) | Fifth wave (n) | Death cases (n) | Survival cases (n) |
|---|---|---|---|---|---|---|---|---|---|---|
| HA | Favors mammalian adaptation | G186V | V | 4 | 10 | 8 | 3 | 36 | 32 | 29 |
| Receptor-binding site | Q226L | L | 4 | 10 | 5 | 3 | 36 | 31 | 27 | |
| Q226I | I | 0 | 0 | 3 | 0 | 0 | 1 | 2 | ||
| Alter the receptor specificity | S138A | A | 4 | 10 | 5 | 3 | 36 | 32 | 29 | |
| T160A | T | 0 | 0 | 0 | 0 | 1 | 0 | 1 | ||
| T160A | A | 4 | 10 | 5 | 3 | 35 | 32 | 28 | ||
| K193T | K | 4 | 10 | 8 | 3 | 36 | 32 | 29 | ||
| N224K | N | 4 | 10 | 8 | 3 | 36 | 32 | 29 | ||
| G228S | G | 4 | 10 | 8 | 3 | 36 | 32 | 29 | ||
| Cleavage peptides | PKGR ↓ GLF | 4 | 10 | 8 | 3 | 33 | 32 | 26 | ||
| PRGR ↓ GLF | 0 | 0 | 0 | 0 | 3 | 0 | 3 | |||
| NA | Related to drug resistance | Q136K | Q | 4 | 10 | 8 | 3 | 36 | 32 | 29 |
| R152K | R | 4 | 10 | 8 | 3 | 36 | 32 | 29 | ||
| R292K | R | 4 | 10 | 8 | 3 | 36 | 32 | 29 | ||
| H274Y | H | 4 | 10 | 8 | 3 | 36 | 32 | 29 | ||
| I222M | I | 4 | 10 | 8 | 3 | 36 | 32 | 29 | ||
| PA | Species-associated signature positions | V100A | A | 4 | 6 | 6 | 1 | 19 | 22 | 14 |
| V100A | V | 0 | 4 | 2 | 2 | 17 | 10 | 15 | ||
| K356R | R | 4 | 10 | 8 | 3 | 36 | 32 | 29 | ||
| S409N | N | 4 | 9 | 8 | 3 | 36 | 31 | 29 | ||
| S409N | S | 0 | 1 | 0 | 0 | 0 | 1 | 0 | ||
| Increase polymerase activity in mice | L336M | L | 4 | 10 | 8 | 3 | 36 | 32 | 29 | |
| PB1 | Increased transmission in ferrets | I368V | V | 4 | 10 | 8 | 3 | 35 | 32 | 28 |
| I368V | I | 0 | 0 | 0 | 0 | 1 | 0 | 1 | ||
| Increased replication in mammalian cells | L598P | L | 4 | 10 | 8 | 3 | 36 | 32 | 29 | |
| PB1-F2 | Increased pathogenicity in mice(length) | 99aa | 3 | 10 | 7 | 3 | 32 | 32 | 23 | |
| 52aa | 1 | 0 | 1 | 0 | 4 | 0 | 6 | |||
| Altered virulence and antivirus response in mice | N66S | N | 3 | 10 | 7 | 3 | 32 | 32 | 23 | |
| PB2 | Increased virulence in mice | E627K | K | 4 | 8 | 4 | 2 | 24 | 27 | 15 |
| E627K | E | 0 | 2 | 4 | 1 | 5 | 4 | 8 | ||
| E627V | V | 0 | 0 | 0 | 0 | 7 | 1 | 6 | ||
| Enhanced transmission in guinea pigs | D701N | N | 0 | 0 | 1 | 1 | 3 | 4 | 1 | |
| D | 4 | 10 | 7 | 2 | 33 | 28 | 28 | |||
| Enhance the 627K and 701N function | K526R | K | 4 | 10 | 8 | 2 | 15 | 19 | 20 | |
| K526R | R | 0 | 0 | 0 | 1 | 21 | 13 | 9 | ||
| Restored the polymerase activity | M535L | M | 4 | 1 | 3 | 2 | 15 | 10 | 15 | |
| M535L | L | 0 | 9 | 5 | 1 | 21 | 22 | 14 | ||
| Efficient replication in mammalian and avian cells, and higher virulence in mice | A588V | A | 4 | 10 | 5 | 1 | 21 | 25 | 16 | |
| A588V | V | 0 | 0 | 3 | 2 | 14 | 7 | 12 | ||
| A588I | I | 0 | 0 | 0 | 0 | 1 | 0 | 1 | ||
| NP | Host signature amino acids(Avain to Human) | V33I | V | 4 | 10 | 8 | 3 | 36 | 32 | 29 |
| I109V | I | 4 | 10 | 8 | 3 | 36 | 32 | 29 | ||
| M1 | Altered virulence in mice | N30D | D | 4 | 10 | 8 | 3 | 36 | 32 | 29 |
| T215A | A | 4 | 10 | 8 | 3 | 36 | 32 | 29 | ||
| Impacts growth and transmission in the guinea pig model | P41A | A | 4 | 10 | 8 | 3 | 36 | 32 | 29 | |
| Host signature amino acids (Avain to Human) | V115I | V | 4 | 10 | 8 | 3 | 36 | 32 | 29 | |
| M2 | Reduced susceptibility to licensed anti-influenza medications | S31N | N | 4 | 10 | 8 | 3 | 36 | 32 | 29 |
| NS1 | Altered virulence in mice | P42S | S | 4 | 10 | 8 | 3 | 36 | 32 | 29 |
| D92E | D | 4 | 10 | 8 | 3 | 36 | 32 | 29 | ||
| Altered antiviral response in host | N205S | S | 4 | 10 | 8 | 1 | 32 | 28 | 27 | |
| N205R | R | 0 | 0 | 0 | 2 | 0 | 2 | 0 | ||
| N205G | G | 0 | 0 | 0 | 0 | 4 | 2 | 2 | ||
| G210R | G | 4 | 10 | 8 | 3 | 36 | 32 | 29 | ||
| NS2 | Altered antiviral response in host | T48A | A | 4 | 10 | 8 | 1 | 36 | 30 | 29 |
| T48E | E | 0 | 0 | 0 | 2 | 0 | 2 | 0 | ||
3.3 Reassortment aspects of H7N9 IAVs established in the Anhui province
The H7N9 isolates established from the Anhui province may serve as donors to participate in the reassortment of at least eight strains of IAVs (Table 2). They might be used as major donors to provide several segments, including HA for H7Ny subtype IAVs. Subsequently, two H7N2 and one H7N6 strain were generated. In the strain, A/chicken/Jilin/SD020/2014(H7N2), A/Anhui/01881/2014(H7N9) provided all segments except NA and NS, whereas A/Environment/Anhui/09183/2014(H9N2) offered NS. The NA segments might also be derived from A/Environment/Anhui/09183/2014(H9N2) or other unexamined HxN2 subtypes IAVs. In the process of generating the strain A/duck/Fujian/5.25_FZHX0665-O/2017(H7N2) through reassortment, two H7N9 isolates might donate all segments except NA, where A/Anhui/29176/2017(H7N9) provided PB2, PA, HA, NP, MP, and NS. A/Anhui/13449/2017(H7N9) contributed to the PB1 segment and its NA segment might have originated from some unexamined HxN2 subtype IAVs (most likely H9N2). Similarly, two H7N9 isolates were involved in the reassortment process of A/chicken/Jiangxi/14501/2014(H7N6) and one was from the Anhui province (Figure 5A–C).
| Reassortant | Major donator | Minor donator | Breakpoints | Segment | |
|---|---|---|---|---|---|
| 1 | A/chicken/Jilin/SD020/2014(H7N2) | A/Anhui/01881/2014(H7N9) | A/Environment/Anhui/09183/2014(H9N2) | 9048-end | NEP |
| 2 | A/duck/Fujian/5.25_FZHX0665-O/2017(H7N2) | A/Anhui/29176/2017(H7N9) | A/Anhui/13449/2017(H7N9) | 2490-4644 | PB1 |
| 3 | A/chicken/Jiangxi/14501/2014(H7N6) | A/Anhui/01883/2014(H7N9) | A/Shanghai/02/2013(H7N9) | 4303-7315 | PA |
| 4 | A/chicken/Jiangxi/08.24_NCDZT49X2-OC/2017(H9N9) | A/Shanghai/02/2013(H7N9) | A/Anhui/29177/2017(H7N9) | 1-2339,4930-6583, 9671-10615 | PB2, PA, MP |
| 5 | A/environment/Yunnan/07.11_DQXY20/2015(H5N6) | A/Yunnan/DQ002/2015(H5N6) | A/Anhui/33225/2015(H7N9) | 6774-8176 | NP |
| 6 | A/duck/Jiangxi/05.07_NCJD0027A-O/2015(H6N6) | A/Pigeon/China/R095/2013(H6N2) | A/Anhui/39280/2015(H7N9) | 1-2319, 6750-9184 | PB2, NP, MP |
| 7 | A/duck/Hunan/1.17_YYGKK90-OC/2017(H6N6) | A/Anhui/45322/2016(H7N9) | A/Anhui/29176/2017(H7N9) | 1-2350 | PB2 |
| 8 | A/chicken/Jiangxi/13238/2014(H10N6) | A/Anhui/33227/2015(H7N9) | A/Anhui/29177/2017(H7N9) | 1-2350, 4554-6527 | PB2, PA |
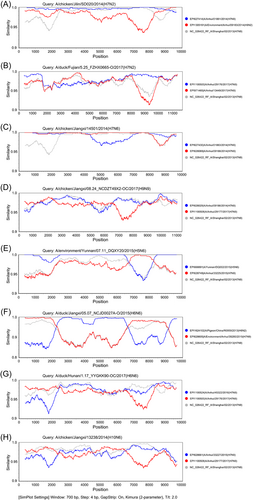
These H7N9 isolates may also be used as major donors to deliver several segments, including NA for HxN9 subtype IAVs. In the strain A/chicken/Jiangxi/08.24_NCDZT49X2-OC/2017(H9N9), A/Shanghai/02/2013(H7N9) offered PB1, NP, NA, and NS segments, whereas A/Anhui/29177/2017(H7N9) provided PB2, PA, MP, and other segments. Unexamined subtypes of H9Ny (most likely an H9N2 strain) provided HA segments for this above-mentioned H9N9 strain (Figure 5D).
These H7N9 viruses also provide one or more segments of the six internal protein-coding genes for several IAVs. For example, A/Anhui/33225/2015(H7N9) provided NP segment for the reassortant A/environment/Yunnan/07.11_DQXY20/2015(H5N6); A/Anhui/39280/2015(H7N9) provided PB2, NP, and MP for A/duck/Jiangxi/05.07_NCJD0027A-O/2015(H6N6); A/Anhui/45322/2016(H7N9); and A/Anhui/29176/2017(H7N9), respectively, provided PB1, PA, NP, MP, and NS, and PB2 segments for A/duck/Hunan/1.17_YYGKK90-OC/2017(H6N6); A/Anhui/33227/2015(H7N9); and A/Anhui/29177/2017(H7N9) provided PB1, NP, MP, and NS, and PB2 and PA segments for A/chicken/Jiangxi/13238/2014(H10N6), respectively (Figure 5E–H).
4 DISCUSSION
In this study, the epidemiological characteristics of outbreak Waves 1–5 of H7N9 influenza in Anhui province are discussed. The phylogenetic features and key amino acid residue substitutions of the 61 H7N9 isolates established in the outbreaks were analyzed. The possibility of these isolates serving as reassortant donors to generate novel IAV subtypes was also explored.
Before September 2018, six waves of H7N9 influenza outbreaks were recorded in China. After no reported human cases for more than a year, the re-emergence of a human HPAI H7N9 virus infection was confirmed in mainland China in late March 2019.35 This implies that the H7N9 IAVs were not eradicated since 2013, although the virus-positive rates substantially decreased from 2016 to 2019 due to an increasing number of provinces having started to close live poultry markets (LPMs)36-38 or take special measures at the human-animal interface to lower the risks of human infection.39 In Anhui province, five outbreak waves were observed before 2018, which was consistent with nationwide epidemics.40 However, the sixth epidemic wave did not appear on schedule in Anhui. Since Anhui province is located in the YRD, which was one of the first places to be hit by the H7N9 outbreak with the first reported human infection,8 this abnormal phenomenon indicates that the H7N9 epidemic in Anhui may be different from that elsewhere in the country. Thus, its future trend cannot be ignored.
Our study revealed that 61 H7N9 isolates established from the Anhui province might participate in the generation of at least eight other IAV subtypes and their clusters through gene reassortment. Reassortment occurs only when the host is simultaneously infected (coinfected) by two or more viruses.11, 12 The relatively low virulence or pathogenicity of viruses is the premise of co-infection. When the virulence of a virus is enough to kill the host, the coinfected viruses cannot maintain basic replication, proliferation, or gene reassortment.11, 12, 14-16 In some classical low pathogenic IAV subtypes such as H1N1, H1N2, and H3N2, a large number of infections provide chances for coinfection by two or more subtypes. Thus, reassortment events are very common. This often results in global epidemics and even pandemics.41-43 In our study, all the 61 H7N9 isolates established in Anhui province were characterized by low pathogenicity in poultry with only a single amino acid “R” or “K” in the cleavage site of HA protein, making it possible to coinfect a host simultaneously.4, 5 The existence of impure isolates with different subtypes has been reported in many studies.44, 45 In the latest nationwide AIV surveillance, Bi et al. unveiled that AIVs (mainly the subtypes of H9N2, H7N9, and H7Ny, H5N1, and other H5Ny) isolation rates in the YRD and the South-Central regions of China were higher than 20.00% and that coinfection or “impure” AIVs led to antigenic or subtype shift by way of reassortment.39 All the results highlight the necessity of constant surveillance of AIV in LPMs.
The characteristic of adaptability to humans of the 61 H7N9 isolates established from Anhui province in our study is a risk that cannot be ignored, whether encoded by the external proteins HA and NA or the six internal proteins encoded by the PB2, PB1, PA, NP, MP, and NS genes. Characteristic sites such as PB2-535L, PB2-588V, PB1-368V, PA-100A, M1-30D, M1-41A, and M1-215A, and amino acid residue substitutions such as PB2-K526R, PB2-E627K, D701N, PA-K356R, PA-S409N, HA-S138A, HA-T160A, HA-G186V, and HA-Q226L/I were found in all or part of the 61 H7N9 isolates established from the five outbreaks in Anhui province. These sites are related to mammalian tropism,46-52 which can increase the binding of human or other mammalian cell receptors to the virus. Therefore, despite their lower pathogenicity, these H7N9 IAVs showed increased abilities and risk of infecting humans, which deserves closer attention. This implies that AIV generated by the reassortment of such viruses is likely to become a novel subtype to infect humans and cause spillover infections or even more serious epidemics in the future. Therefore, as long as the low pathogenic H7N9, H9N2, and H5N1 persist, the emergence of novel AIVs that can cause human epidemics in the YRD and South-Central regions of China may be only a matter of time. Based on this study and other results,53-57 we propose that H7N9, H9N2, and H5N1 can serve as stable and persistent gene pools for IAVs in the YRD and South-Central regions of China. Under these conditions, novel AIV subtypes may be continuously generated by reassortment and can subsequently cause human epidemics through accidental spillover infections (Figure 6).
In summary, our study indicated that the 61 H7N9 viruses established from five outbreak waves in the Anhui province have low pathogenicity and high human or mammalian tropism. In long-term circulation and transmission, these H7N9 isolates have introduced novel AIV subtypes such as H7N2, H7N6, H9N9, H5N6, H6N6, and H10N6 through gene reassortment by donating one or more segments of HA, NA, and six internal protein-coding genes. The novel AIVs may have obtained human-type receptor-binding abilities from their donors and prefer binding to them. Considering the continual threat of emerging avian influenza in the future, constant monitoring of AIVs should be conducted more closely for agricultural and public health.
AUTHOR CONTRIBUTIONS
Conceptualization by Yihan Lu, Jiabing Wu, Jun He, and Sai Hou. Methodology by Chenglong Xiong, Jun He, and Sai Hou. Formal analysis by Jun He, Sai Hou, Chenglong Xiong, Xiaoyu Zhou, and Linjie Hu. Experiment by Jun He, Sai Hou, Lei Gong, Junling Yu, Xiaoyu Zhou, Qingqing Chen, Yuan Yuan, Lan He, Linjie Hu, Meng Zhu, Weiwei Li, Yonglin Shi, Yong Sun, Haifeng Pan, and Bin Su. Resources by Yihan Lu, and Jiabing Wu. Writing – original draft by Jun He, Sai Hou, Jiabing Wu, Lei Gong, Junling Yu, Xiaoyu Zhou, Qingqing Chen, Yuan Yuan, Lan He, Linjie Hu, Meng Zhu, Weiwei Li, Yonglin Shi, Yong Sun, Haifeng Pan, and Bin Su. Writing – review and editing by Chenglong Xiong, Yihan Lu, and Jiabing Wu. Supervision by Yihan Lu, Jiabing Wu, and Chenglong Xiong. Project administration by Yihan Lu, Jiabing Wu, and Chenglong Xiong; Funding acquisition by Yihan Lu, Jiabing Wu, and Chenglong Xiong.
ACKNOWLEDGMENTS
The authors acknowledge Prof Weifeng Shi and Juan Li for kindly providing us with suggestions during the writing and revision of the manuscript. Professors Shi and Li are research professors at the School of Public Health, Shandong First Medical University. This research was supported by the National Natural Science Foundation of China (81872673), the Three-Year Action Plan of Shanghai Public Health System Construction-Key Discipline Construction, 2020–2022 (GWV-10.1-XK03), the Scientific Research Projects of Health Commission of Anhui Province in 2021 (AHWJ2021a030), and the Open Funding of the State Key Laboratory for Diagnosis and Treatment of Infectious Diseases and the First Affiliated Hospital, Zhejiang University School of Medicine (SKLID2021KF04). The funders had no role in the study design, data collection, analysis, interpretation, or writing of the report. The corresponding author has full access to all study data and the final responsibility for the decision to submit for publication.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
The study involved the use of existing sequence data in the GISAID and GenBank databases. All data included in this study had no identifiers of humans or animals.
Open Research
DATA AVAILABILITY STATEMENT
The datasets analyzed in this study, both from us and other researchers, are available in the Global Initiative on Sharing All Influenza Data (GISAID) EpiFlu at https://www.gisaid.org and the National Center for Biotechnology Information (NCBI) Flu databases at https://www.ncbi.nlm.nih.gov/. We acknowledge the two databases for allowing us to use these sequences freely and conveniently.




