Do CCR5 (CCR5Δ32) and TLR3 (RS5743313) gene polymorphisms prevent chronic hepatitis B infection?
Abstract
Hepatitis B virus (HBV) is still a significant health problem in human. HBV severity or sensitivity of patients may be based on the individual genetic factors significantly. The aim of this study is to investigate the association of CCR5 (CCR5Δ32), TLR3 (rs5743313) functional gene polymorphisms, interferon-gamma (IFN-ɣ) level in HBV infection, which are thought to play an important role in innate and acquired immunity in patients who have undergone HBV seroconversion and those who have chronic hepatitis B disease and receive treatment. One hundred patients who are became naturally immune against HBV infection (HBsAg negative, anti-HBc IgG, and anti-HBs IgG positive), and 100 patients with chronic hepatitis B infection (>6 months HBsAg positive) who are receiving oral antiviral therapy were compared for CCR5Δ32, TLR3 (rs5743313) genotypes and serum IFN-ɣ level. It was found that CCR5Δ32 polymorphism (Wt/Δ32 and Δ32/Δ32) was significantly higher in the chronic hepatitis B group (p = 0.048) but not for TLR3 gene polymorphism. However, serum IFN-ɣ level was significantly higher in the HBV seroconversion group (75 ± 89 ng/ml) than in the chronic hepatitis B group (4.35 ± 17.27 ng/ml) (p < 0.001). In conclusion, a higher CCR5Δ32 allele frequency in patients with chronic hepatitis B might be considered as a marker of progression to chronic hepatitis.
1 INTRODUCTION
Hepatitis B virus (HBV), which is in the Hepadnaviridae family, still continues to be an important health problem due to a number of liver diseases such as acute and chronic hepatitis, cirrhosis, hepatocellular cancer (HCC).1-3 With the integration of HBV into the host genome, nonfunctional T cell responses and deleterious immunopathology are triggered.4 Although the main mechanisms responsible for the natural course of HBV infection are not clear, the genetic factors of infected individuals play a crucial role in the immunopathogenesis of HBV infection.5, 6 Studies show that especially single nucleotide polymorphisms (SNP; substitution of one nucleotide with another nucleotide) in various immune cytokines and receptor genes play an important role in immunity, clinical course, and recovery from HBV infection.7, 8
CC chemokine receptor 5 (CCR5) is expressed on the surface of natural killer (NK) cells, CD8+ T lymphocytes, and macrophages, and plays an important role in the migration of interferon-gamma (IFN-ɣ)-producing CD4+, CD8+, and NK cells to the liver after inflammation in chemotaxis and naturally throughout the virus clearance process.9, 10 In addition, CCR5 can contribute to the immunopathology of inflammation by taking part in the regulatory and memory T cell responses.11 CCR5 function is altered by various polymorphisms, one of which is a 32-base pair (bp) deletion (Δ32) that causes a frameshift in the coding sequence of the receptor's second extracellular loop, resulting in loss of function of the prematurely truncated protein.12 CCR5Δ32 polymorphism is found to be related for resistance or susceptibility to various viral diseases, such as hepatitis C, human immunodeficiency virus, influenza viruses.13-15 Studies indicated that CCR5Δ32 polymorphism also can be involved in clinical outcomes, severity of hepatitis, and resistance/protective effect to HBV infection in different populations in many countries.16-18 However, there is no scientific data about the relation of CCR5Δ32 polymorphism and clinical outcome of HBV infection in Turkish population.
Toll-like receptors (TLRs), one of the pathogen recognition receptors, recognize pathogen-associated molecular structures and activate innate immune mechanisms against pathogens.19, 20 TLR3, a member of the TLR family, has been shown to have a role in the recognition of HBV.21 A defective or decreased TLR3 response can lead to a weakening of the immune response against HBV.22 It is reported that a number of TLR3 polymorphisms including rs5743313 are associated with severity of some viral diseases, that is, enteroviruses, influenza viruses, and HBV by reducing the efficiency of the virus recognition mechanisms.23-25 TLR3-rs5743313 is an intronic polymorphism, but is located near exon 4 which encodes transmembrane signal induction region of the receptor. It is thought to play an essential role in receptor function by reducing the efficiency of the virus recognition mechanisms.23 A study showed that TLR polymorphisms including rs5743313 is significantly associated with Saudi Arabian chronic hepatitis B (CHB) patients.25 However, relation between TLR3 rs5743313 polymorphism and clinical outcome of hepatitis B has not been investigated in Turkish population.
Non-cytopathic HBV-induced liver damage and viral control are regulated by the immune system.26, 27 The host coordinates the response against inflammatory diseases by regulating the proliferation, differentiation, and function of immune cells with the antiviral cytokines it produces to limit HBV infection.26-28 It is well known that cytokines such as IFN-ɣ and TNF-alpha play a central role in the clearance of acute infection or the persistence of HBV.29 Serum IFN-ɣ levels are found to be higher in CHB patients in comparison to healthy controls and increased serum levels of IFN-γ are correlated with HBV-related active hepatitis.30, 31 To date, there is no study for investigation for relation of serum IFN-ɣ levels of hepatitis B patients and CCR5Δ32 and TLR3 (rs5743313) polymorphisms by comparison the immune and CHB groups.
The aim of our study was to detect the association of CCR5 (CCR5Δ32) and TLR3 (rs5743313) functional gene polymorphisms and serum IFN-ɣ level in patients infected with HBV who are subsequently seroconverted (immune patients) and in patients receiving treatment for CHB disease (CHB patients). Due to lack of knowledge of the role of CCR5 (CCR5Δ32) and TLR3 (rs5743313) polymorphisms in HBV infected Turkish population, this study aimed to investigate the association of these polymorphisms with clinical course of HBV infection in Turkey.
2 MATERIALS AND METHODS
2.1 Patients
Two hundred patients who applied to Kırıkkale University Faculty of Medicine Infectious Diseases and Clinical Microbiology Outpatient Clinic and have had hepatitis B infection were included in the study. Patients were divided into two groups: (i) “immune group” which is consisted of 100 patients who had been infected by HBV and became immune (HBsAg negative, anti-HBc IgG, and anti-HBs IgG positive) and (ii) “chronic hepatitis B group” which is consisted of 100 patients who were diagnosed with CHB (HBsAg positive for at least 6 months) and received nucleos(t)ide analogue therapy (tenofovir disoproxil fumarate in 68 patients; tenofovir alafenamide fumarate in 7 patients; entecavir in 17 patients; lamivudine in 8 patients). Serum and whole blood samples were obtained from the volunteers included in the study and stored at −20°C for further analyses.
2.2 DNA extraction
Genomic DNA extraction from whole blood samples was performed using Exgene Blood, Clinic, Cell SV mini (Ver 4.1) kit (GeneAll Biotechnology) according to the manufacturer's recommendations. DNA samples were stored at −20°C until further use in conventional PCR and real-time PCR.
2.3 CCR5Δ32 genotyping by conventional PCR
CCR5Δ32 genotyping in patients were investigated by conventional PCR. Total of 50 µl mixture containing DNA, 10× Taq Buffer, 1.25 mM dNTP (R0181; Thermo Scientific Fisher), 10 pmol forward and reverse primers (Ella Biotech) (Tables 1), 5 U/µl Taq DNA polymerase (MG-KTAQ-01; Hibrigen), and sterile distilled water was prepared. The reaction conditions for PCR were as follows: 94°C for 3 min; 35 cycles of 94°C for 40 s, 58°C for 40 s, 72°C for 40 s, and the final elongation step at 72°C for 10 min. Visualization of the PCR product was carried under UV light following electrophoresis on 3% agarose gel stained with ethidium bromide (Illuminyx; NYXTechnik). Sterile distilled water was used as negative control and positivity of samples are confirmed by Sanger sequencing (Sentebiolab).
| Primer | Sequence (5′→3′) | Product size (bp)a | References |
|---|---|---|---|
| CCR5-F | TCAAAAAGAAGGTCTTCATTACACC | 241 | [32] |
| CCR5-R | AGCCCAGAAGAGAAAATAAACAATC | 209 | |
| Primer/probe | |||
| TLR3-F | CATTGGTGTCATCCTCCTGAGA | ||
| TLR3-R | GCAGGGCGGCAGAGT | 71 | [23] |
| TLR3-P1b | FAM-TCTCCCGACCTCTCC-BHQ1 | ||
| TLR3-P2c | HEX-TCTCCCAACCTCTCC-BHQ1 |
- Abbreviations: bp, base pair; CCR5, CC chemokine receptor 5; TLR, toll-like receptors.
- a In CCR5Δ32 genotyping, as a result of PCR, wild type gene product was obtained as 241 bp single band, heterozygous deletion gene product as 241 and 209 bp two bands, and homozygous deletion gene product as 209 bp single band.
- b TLR3-P1 (FAM) probe determines wild type.
- c TLR3-P2 (HEX) probe determines mutated genotype.
2.4 TLR-3 genotyping by real-time PCR
TLR-3 (rs5743313) genotyping of patients was performed by real-time PCR. Reactions were set up with 1 µl DNA, 2× Probes master mix (P02-01-05; ABT Biotechnology), 10 pmol/µl forward and reverse primer (final concentration 0.5 pmol/µl), 10 pmol/µl probe (final concentration 0.25 pmol/µl each) (Table 1), and ultrapure water (809-115-CL; Wisent) in a total volume of 20 µl. The amplification was carried out under the following conditions: 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 15 s, 55°C for 20 s, 72°C for 20 s, in LightCycler 96 real-time PCR instrument (Roche) and genotyping results were evaluated using LightCycler 96 SW1.1 software (Roche). TLR3 gene polymorphism; FAM + /HEX− was genotyped as wild type, FAM + /HEX+ as heterozygous mutant, and FAM-/HEX+ as homozygous mutant.
2.5 Measurement of IFN-ɣ level
Measurement of IFN-ɣ level in serum samples was performed using the Human Interferon Gamma, IFN-G ELISA Kit (Bioassay Technology Laboratory) using the manufacturer's recommended protocol.
2.6 Immunoassay for serological markers
Serum HBsAg, HBeAg, anti-HBe, anti-HBc, and anti-HBs IgG was quantified using the Architect Reagent Kit (Abbott), according to the manufacturer's instructions.
2.7 Biochemical parameters
Serum aspartate transferase (AST), alanine transferase (ALT) levels were determined using the Roche cobas c 702 systems and alpha-fetoprotein were analyzed by Roche cobas e 801 systems.
2.8 HBV DNA viral load
HBV DNA viral load testing was performed using the artus HBV QS-RGQ Kit (Qiagen) according to the manufacturer's instructions and LightCycler 480 real-time PCR instrument (Roche).
2.9 Statistical analysis
Statistical analyzes were performed using SPSS version 20.0 software. Descriptive statistics; were given as numbers and percentages for categorical variables, and as mean ± standard deviation for numerical variables. The conformity of the variables to the normal distribution was examined using the histogram graph and the Kolmogorov−Smirnov test. The Mann−Whitney U test was used for comparisons between the two groups for numerical variables that did not show normal distribution. χ2 test was used for comparisons between categorical variables and for the deviation of Hardy−Weinberg equilibrium (HWE). In the results, a p value below 0.05 was considered statistically significant. Odds ratio (OR) was calculated by MedCalc Software Ltd. OR calculator33 and HWE was calculated by Excel (Microsoft Office).
3 RESULTS
3.1 Demographic, clinical features, and biochemical profile of the patients by groups
The age, gender distribution of the patients, clinical feature, biochemical profile (AST, ALT) among the groups are shown in Table 2. Due to lack of some data of the patients in immune group because they have not applied to our clinic in acute phase of HBV infection, HBV DNA load has not been checked in immune group.
| Patient groups | ||
|---|---|---|
| Features | Immune group | Chronic hepatitis B group |
| Number of patients | 100 | 100 |
| M/F gender ratio | 45/55 | 51/49 |
| Age (years, arithmetic mean ± standard deviation) | 60.35 ± 15.02 | 51.14 ± 12.40 |
| M/F age ratio | 60.13/60.53 | 52.67/49.55 |
| Smoking, n (%) | 36 (36) | 35 (35) |
| Laboratory parameters (mean ± SD) | ||
| AST (IU/L) | 22.93 ± 11.56 | 20.85 ± 8.32a |
| ALT (IU/L) | 23.01 ± 14.84 | 21.21 ± 12.59a |
| AFP (ng/ml) | No data | 3.03 ± 2.69 |
| HBV-DNA, log10 (copy/ml)b | No data | 8.93 ± 9.78 |
- Abbreviations: AFP, alpha-fetoprotein; ALT, alanine transferase; AST, aspartate transferase; HBV, hepatitis B virus.
- a Posttreatment ALT-AST.
- b Pretreatment HBV DNA.
3.2 CCR5Δ32 polymorphism and TLR3 gene rs5743313 polymorphism results
For the CCR5Δ32 deletion Wt/Wt genotype was defined as homozygosity for the CCR5 wild-type allele; the Wt/Δ32 genotype was defined as heterozygosity for the CCR5 wild-type allele and the CCR5Δ32 deletion; the Δ32/Δ32 genotype was defined as homozygosity for CCR5Δ32 deletion (Figure 1). The genotype results of conventional PCR were confirmed by Sanger sequencing (data not shown). Wt/Wt genotype was detected in 95 out of 100 patients and 87 out of 100 patients in immune group and CHB group, respectively. Homozygous Δ32 genotype was detected only in 1 patient in CHB group (Table 3). The genotype distribution was in HWE (Wt/Wt 182 observed [O], 181.5 expected [E]; Wt/Δ32 17 O, 18.1 E; Δ32/Δ32 1 O, 0.5 E).
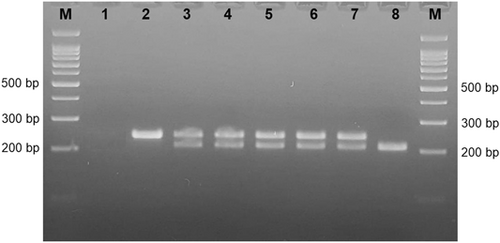
| CCR5 Δ32 | Immune group | Chronic hepatitis B group | p Value | TLR3 rs5743313 | Immune group | Chronic hepatitis B group | p Value |
|---|---|---|---|---|---|---|---|
| Wt/Wt | 95 | 87 | 0.048a | CC | 62 | 56 | 0.388a |
| Wt/Δ32 | 5 | 12 | 0.076a | CT | 34 | 39 | 0.463a |
| Δ32/Δ32 | 0 | 1 | - | TT | 4 | 5 | 0.733a |
| Wt/Δ32 + Δ32/Δ32 versus Wt/Wt | 5−95 | 13−87 | 0.048a | CT + TT versus CC | 38−62 | 44−56 | 0.388a |
| Δ32/Δ32 versus Wt/Δ32 + Wt/Wt | 0−100 | 1−99 | - | TT versus CT + CC | 4−96 | 5−95 | 0.733a |
| Total | |||||||
| Wt allele frequency | 0.975 | 0.93 | 0.088a | C allele frequency | 0.79 | 0.755 | 0.611a |
| Δ32 allele frequency | 0.025 | 0.07 | 0.194a | T allele frequency | 0.21 | 0.245 | 0.502a |
| OR; (95% CI) | |||||||
| Wt/Δ32 + Δ32/Δ32 versus Wt/Wt | 2.839; (0.972−8.290) | 0.056 | CT + TT versus CC | 1.282; (0.728−2.255) | 0.388 | ||
| Δ32/Δ32 versus Wt/Wt + Wt/Δ32 | - | - | TT versus CT + CC | 1.263; (0.329−4.848) | 0.733 | ||
| Δ32 versus Wt allele frequency | 2.936; (1.037−8.311) | 0.043 | T versus C allele frequency | 1.221; (0.764−1.951) | 0.4 | ||
- Note: p value < 0.05 was considered statistically significant as bold.
- Abbreviations: CCR5, CC chemokine receptor 5; CI, confidence intervals; OR, odd ratio; TLR, toll-like receptors.
- a χ2 test.
Wt/Wt genotype was statistically higher in immune group compared to CHB group (p = 0.048, χ2 test) for CCR5Δ32 polymorhism. There was no statistically significant difference between the groups in Wt/Δ32 genotype (p = 0.076, χ2 test). The total incidence of Wt/Δ32 and Δ32/Δ32 genotypes was 5% in immune group and 13% in CHB group, and this difference was statistically significant (p = 0.048, χ2 test). The OR with 95% confidence intervals (OR, 95% CI) were calculated for each polymorphism using three genetic association models, including dominant, recessive, and allele models. There was no statistically significant difference between two groups for both polymorphisms, except; Δ32 allele frequency was statistically higher in immune group compared to CHB group (OR: 2.963, 95% CI: 1.037−8.311; p = 0.043) (Table 3).
For TLR3 gene rs5743313 polymorphism, CC, CT, and TT genotypes, including C wild type allele and T variant allele, were determined (Figure 2). Genotype and allele frequencies of TLR3 rs5743313 in immune and chronic hepatitis groups are shown in Table 3. For TLR3 gene rs5743313 polymorphism no statistically significant difference was found when CT, TT, and CC genotypes were compared for both groups. The genotype distribution was in HWE (p = 0.587).
There was a higher rate of the CCR5Δ32 Wt/Wt genotype in the HBeAg negative patients compared to the HBeAg positive patients (p = 0.007). There was no statistically significant difference between the viral load, ALT and AST levels, histological activity index (HAI), and fibrosis score with CCR5Δ32 and TLR3 rs5743313 genotypic polymorphisms distribution in patients of CHB group (Table 4).
| Genotypes of polymorphism | HBeAg positive | HBeAg negative | p Value | Viral load ≤105 copies/mla | Viral load >105 copies/mla | p Value | |
|---|---|---|---|---|---|---|---|
| CCR5 | Wt/Wt | 8 | 79 | 0.007* | 30 | 57 | 0.362* |
| Wt/Δ32 | 2 | 10 | 5 | 7 | |||
| Δ32/Δ32 | 1 | - | 1 | - | |||
| TLR3 | CC | 8 | 48 | 0.541* | 22 | 34 | 0.684* |
| CT | 3 | 36 | 12 | 27 | |||
| TT | - | 5 | 2 | 3 |
| HAI ≤ 5 | HAI > 5 | Fibrosis ≤ 2 | Fibrosis > 2 | ||||
|---|---|---|---|---|---|---|---|
| CCR5 | Wt/Wt | 24 | 63 | 0.819* | 63 | 24 | 0.244* |
| Wt/Δ32 | 2 | 10 | 9 | 3 | |||
| Δ32/Δ32 | 1 | - | 1 | - | |||
| TLR3 | CC | 11 | 45 | 0.244* | 37 | 19 | 0.393* |
| CT | 12 | 27 | 31 | 8 | |||
| TT | 2 | 3 | 3 | 2 |
| ALT ≤ 40 IU/Lb | ALT > 40 IU/Lb | AST ≤ 40 IU/Lb | AST > 40 IU/Lb | ||||
|---|---|---|---|---|---|---|---|
| CCR5 | Wt/Wt | 52 | 35 | 0.067* | 58 | 29 | 0.402* |
| Wt/Δ32 | 8 | 4 | 10 | 2 | |||
| Δ32/Δ32 | 1 | - | 1 | - | |||
| TLR3 | CC | 38 | 18 | 0.149* | 37 | 19 | 0.723* |
| CT | 19 | 20 | 28 | 11 | |||
| TT | 4 | 1 | 4 | 1 |
- Abbreviations: ALT, alanine transferase; AST, aspartate transferase; CCR5, CC chemokine receptor 5; HAI, histological activity index; HBV, hepatitis B virus; TLR, toll-like receptors.
- a Pretreatment HBV DNA.
- b Pretreatment ALT-AST.
- * χ2 test.
3.3 IFN-ɣ results
Statistical evaluation of IFN-ɣ levels and genotype distribution by groups is given in Table 5. IFN-ɣ levels were significantly higher in immune group compared to CHB group (p < 0.001, Mann−Whitney U test). In both groups, there was no statistically significant difference between the mean IFN-ɣ levels and genotypic distributions.
IFN-ɣ levels (IU/ml) (mean ± standard deviation) |
|||
|---|---|---|---|
| CCR5 Δ32 polymorphism | Δ32/Δ32 and Wt/Δ32 | Wt/Wt | pa Value |
| Immune group | 76.09 ± 70.50 | 75.05 ± 90.17 | 0.710 |
| Chronic hepatitis B group | 68.71 ± 74.86 | 79.02 ± 97.03 | 0.986 |
| TLR3_rs5743313 polymorphism | CT and TT | CC | |
| Immune group | 68.71 ± 74.86 | 79.02 ± 97.03 | 0.986 |
| Chronic hepatitis B group | 2.33 ± 7.87 | 5.93 ± 21.98 | 0.490 |
| Total | Immune group | Chronic hepatitis B group | |
| 75.10 ± 89.00 | 4.35 ± 17.20 | <0.001 | |
- Abbreviations: CCR5, CC chemokine receptor 5; IFN-ɣ, interferon-gamma; TLR, toll-like receptors.
- a Mann−Whitney U test. p value below 0.05 was considered statistically significant as bold.
4 DISCUSSION
In the clinical course of HBV infection, spontaneous clearance of HBV occurs in most of the HBV-infected patients, while the disease becomes chronic in some patients and causes severe liver damage.2, 3 This variable pattern of infection is thought to be due to environmental, virological, immunological, and host genetic factors. Although the effects of virological and immunological factors are well known, the effects of host genetic factors on HBV infection are not clearly understood.7 In this present study, CCR5Δ32 and TLR3 rs5743313 polymorphisms are investigated throughout the two different hepatitis B patient groups: one is spontaneously healed and became immune against HBV infection, and the other is CHB patients who are receiving antiviral therapy.
CCR5 plays a critical role in regulating T cell functions by mediating the migration and activation of antiviral type 1 cytokine-secreting T helper and cytotoxic T cells. It is thought that the inability of IFN-ɣ secreting T cells to function fully as a result of the CCR5Δ32 polymorphism, which leads to a decrease in the functional CCR5 receptor, increases the risk of CHB and C diseases.27, 34 In our study, CCR5Δ32 homozygosity was found only in 1 patient in CHB group, while CCR5Δ32 homozygosity and heterozygosity were found to be 5% in immune group and 13% in CHB group, and this difference was statistically significant (p = 0.048) and Δ32 allele frequency was higher than Wt allele frequency in chronic hepatitis group (OR: 2.936; 95% CI: 1.037−8.311) (Table 3). We can speculate that in our cohort, patients who had CCR5Δ32 allele has approximately three times the odds of having chronic HBV infection.
The significant elevation of the CCR5Δ32 polymorphism in CHB group supports the hypothesis that the CCR5 receptor cannot fully function and may increase the risk of CHB disease. Suneetha et al.16 reported that patients with CHB disease and healthy individuals were compared and CCR5Δ32 heterozygosity was found to be higher in patients with CHB compared to healthy controls (4.2% vs. 0.73%, p = 0.05), and CCR5Δ32 heterozygosity was associated with susceptibility to HBV infection. In some studies, however, such a relationship could not be demonstrated, possibly due to the very low frequency of CCR5Δ32 polymorphism in the ethnic populations studied.18, 35, 36 On the contrary to these findings, Thio et al.37 reported that the CCR5Δ32 allele was detected more frequently in those who recovered from the infection compared to those with CHB infection (13% vs. 7.4%), and it was found that the CCR5Δ32 polymorphism reduced the risk of developing CHB infection by half. Abdolmohammadi et al.17 showed that CCR5Δ32 polymorphism was higher in healthy controls than in HBV-infected patients, and it was stated that CCR5Δ32 polymorphism may have a protective effect against HBV infection, at least in the Iranian population.
Because the frequency of CCR5Δ32 polymorphism is highly variable among different populations, the effect of this polymorphism in one population may not be valid for other populations.11 In 1.3 million stem cell donors from 87 countries where the CCR5Δ32 polymorphism was investigated, the CCR5-Δ32 allele was found to be more common in European countries (e.g., 16% in Norway, 11% in Germany), and lower in Asian and African countries. In the same study, the frequency of CCR5-Δ32 allele was found to be 3.4% in 36 036 individuals in Türkiye.38
Various factors such as the inhibitory effects of HBV, genetic variations, and epigenetic factors have been reported to affect TLR3 expression.22 SNPs in the TLR3 gene are thought to be among the factors affecting susceptibility to viral pathogens, including HBV.39 In our study, assuming that TLR3 gene rs5743313 SNP may be associated with recovery from HBV infection or becoming chronic, we determined three genotypes for TLR3 gene rs5743313 polymorphism, namely CC, CT, and TT. However, we did not detect any significant difference between the groups (Table 4). We thought that TLR3 gene rs5743313 polymorphism had no significant effect on hepatitis B infection. Contrary to our study, some SNPs in the TLR3 gene have been associated with susceptibility to various diseases, including CHB and HCC. For example, in the study of Al-Qahtani et al.,25 the relationship of 9 SNPs in the TLR3 gene with HBV infection was examined, and when haplotype analysis was performed, it was stated that the GCGA haplotype in 4 SNPs (rs5743313, rs5743314, rs5743315, rs1879026) had a significant effect on susceptibility to HBV infection. It was thought that the rs1879026 polymorphism may also have a protective role against the development of CHB infection. In the study of Ezzel-Din et al.,40 the distribution of 6 SNPs, including rs5743313, in the TLR3 gene was examined, and GCTCCA and CCA haplotype frequencies were found to be significantly higher in the group with HBV clearance compared to CHB patients, and it was stated that these polymorphisms may have a protective role against HBV infection. In the study of Huang et al.,41 it was shown that TLR3 gene rs3775290 polymorphism may be a protective factor for CHB disease, HBV-associated liver cirrhosis and HCC. In the study of Fischer et al.,42 it was determined that the risk of developing CHB disease increased in the presence of TLR3 gene rs3775291 and rs5743305 polymorphisms. In the study of Chen et al.,43 TLR3 gene rs377529 polymorphism was shown to be associated with a significant decrease in the risk of HBV-related HCC. In the study of Rong et al.,19 it was stated that the TLR3 gene C234T polymorphism is associated with a predisposition to CHB.
One can speculate that larger samples should be studied to fully understand the effect of TLR3 gene rs5743313 polymorphism and other TLR3 gene SNPs on HBV infection. Studies in different stages of HBV infection and in different ethnic populations are important in understanding this.
IFN-ɣ is produced from innate (e.g., NK cells, NKT cells, macrophages, and myelomonocytic cells) and adaptive (e.g., TH1 cells, cytotoxic T lymphocytes, and B cells) immune system cells.44 Resolution of HBV infections is dependent on the killing of infected cells by viral antigen-specific cytotoxic T lymphocytes, and most likely interferons, including IFN-ɣ, and proinflammatory cytokines such as TNF-α, inhibiting viral replication by non-cytolytic pathways.45 In our study, serum IFN-ɣ level was significantly higher in immune group patients (mean 75 ± 89 ng/ml) compared to CHB group patients (mean 4.35 ± 17.27 ng/ml) (p < 0.001) (Table 5). In two separate studies, IFN-ɣ levels were found to be higher in patients with HBsAg spontaneous seroconversion than in patients with CHB.46, 47 In the study of Chu et al.,31 IFN-ɣ level was found to be significantly higher in acute hepatitis B patients compared to CHB patients, and it was thought that this may be related to the sensitization of cytotoxic T lymphocytes by HBV antigens and then induction of IFN-ɣ production. Our results, which are consistent with these studies in which the IFN-ɣ level was found to be higher in patients with HBsAg seroconversion compared to the patients in the CHB group, suggests that IFN-ɣ is secreted from CD8+ T cells and helps to ensure clearance of the virus in acute hepatitis B recovery.
In conclusion, the higher CCR5Δ32 allele frequency in patients with CHB was considered as a sign of progression to chronic hepatitis. However, it is clear that disease stage, ethnicity, and many other determinants are also important in disease progression. Studies on larger populations with different polymorphisms in different regions are needed to clearly determine the effects of CCR5 and TLR3 genes and IFN-ɣ level on the severity of hepatitis B infection.
AUTHOR CONTRIBUTIONS
Burçin Tuncel, Emel Aksoy, and Ahmet Kürşat Azkur performed the experiments. Burçin Tuncel, Sedat Kaygusuz, Derya Beyza Sayın, Emel Aksoy, and Ahmet Kürşat Azkur analyzed the data, interpreted the results, and wrote the manuscript. Derya Beyza Sayın, Ahmet Kürşat Azkur, Burçin Tuncel, and Sedat Kaygusuz designed this study.
ACKNOWLEDGMENT
This work was supported by Scientific Research Projects Coordination Unit of Kırıkkale University, project number 2021/072.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
This study was approved by Kırıkkale University Clinical Research Ethics Committee with the decision dated 15.04.2021 and numbered 2021/04.




