Comparison of secondary attack rate and viable virus shedding between patients with SARS-CoV-2 Delta and Omicron variants: A prospective cohort study
Sung-Woon Kang, Ji Yeun Kim, and Heedo Park contributed equally to this work.
[Correction added on 21 January 2023, after first online publication: the grant number funded by the Ministry of Health & Welfare, Republic of Korea, has been corrected.]
Abstract
There are limited data comparing the transmission rates and kinetics of viable virus shedding of the Omicron variant to those of the Delta variant. We compared these rates in hospitalized patients infected with Delta and Omicron variants. We prospectively enrolled adult patients with COVID-19 admitted to a tertiary care hospital in South Korea between September 2021 and May 2022. Secondary attack rates were calculated by epidemiologic investigation, and daily saliva samples were collected to evaluate viral shedding kinetics. Genomic and subgenomic SARS-CoV-2 RNA was measured by PCR, and virus culture was performed from daily saliva samples. A total of 88 patients with COVID-19 who agreed to daily sampling and were interviewed, were included. Of the 88 patients, 48 (59%) were infected with Delta, and 34 (41%) with Omicron; a further 5 patients gave undetectable or inconclusive RNA PCR results and 1 was suspected of being coinfected with both variants. Omicron group had a higher secondary attack rate (31% [38/124] vs. 7% [34/456], p < 0.001). Survival analysis revealed that shorter viable virus shedding period was observed in Omicron variant compared with Delta variant (median 4, IQR [1−7], vs. 8.5 days, IQR [5–12 days], p < 0.001). Multivariable analysis revealed that moderate-to-critical disease severity (HR: 1.96), and immunocompromised status (HR: 2.17) were independent predictors of prolonged viral shedding, whereas completion of initial vaccine series or first booster-vaccinated status (HR: 0.49), and Omicron infection (HR: 0.44) were independently associated with shorter viable virus shedding. Patients with Omicron infections had higher transmission rates but shorter periods of transmissible virus shedding than those with Delta infections.
1 INTRODUCTION
After it was first reported on November 9, 2021, the Omicron variant soon overtook the Delta variant and has been the dominant strain worldwide.1 After the global surge of the Omicron variant, several studies demonstrated that it had higher transmissibility than the Delta variant.2-4 However, there are limited data comparing shedding of the two variants with adjustment for disease severity and vaccination status. Studies of the viral shedding kinetics of outpatients with asymptomatic or mild Delta or Omicron infections revealed comparable shedding durations.5, 6 However, these studies5, 6 were conducted in vaccinated patients without underlying medical conditions, and most of the patients had only mild symptoms or none at all. Although these data provide a useful basis for determining the duration of isolation of healthcare workers—who mostly have no underlying illness and have completed the initial vaccine series—they are of limited generalizability in terms of viable virus shedding kinetics in patients with Delta and Omicron infections, since the latter have variable clinical manifestations depending on age, underlying disease, and vaccination status in the real world.7 Thus, data on the shedding kinetics of the Delta and Omicron variants in hospitalized patients of various severities and underlying diseases according to vaccination status is still lacking.
Furthermore, the Omicron variant clearly has higher transmissibility than the Delta variant. Some studies indicate that viral loads in upper respiratory tract were lower in patients with Omicron variant infection than in those with prior variant infection.8-10 In addition, Omicron variant infection had similar incubation period compared with Delta variant infection.11 These data suggest that high transmissibility may not be due to the viral load itself or the incubation period, but due to more efficient viral replication in upper respiratory tract.9 Furthermore, Omicron variant with numerous mutations in N-terminal domain and receptor binding domains may be superior in terms of immune-evasion.12, 13 Especially, the evasion of humoral immunity is considered as a major factor contributing to the Omicron spread. Therefore, the subsequent subvariants of Omicron (i.e., BA.5 or BQ.1) may take advantage over the precedent Omicron variants (i.e., BA.1. or BA.2) due to antibody evasion, so it is expected that the sequel subvariants will follow this path. Despite these data, there are limited data on viable viral shedding kinetics between Omicron variant and Delta variant infections. We therefore conducted a prospective cohort study evaluating secondary attack rates and virus shedding kinetics together with longitudinal culture-based virus isolation in hospitalized Delta and Omicron patients of varying severity.
2 METHODS
2.1 Study populations
From February 1, 2021 to May 31, 2022, over the period of the Delta and Omicron surge in South Korea, we prospectively enrolled patients over 18 years confirmed as SARS-CoV-2 infection who were admitted and consented to daily saliva sampling at Asan Medical Center, a 2700-bed tertiary hospital in Seoul, South Korea. Patients with known prior histories of COVID-19 infection were excluded. All participants were diagnosed using SARS-CoV-2-specific RT-PCR. All study participants provided written informed consent, and the study protocol was approved by the institutional review board of Asan Medical Center (IRB 2020-0297). The data on virus kinetics in patients with the Delta variant have been published in part.14
2.2 Definitions
All study populations were classified into four subgroups depending to vaccination status—first booster vaccinated, who completed the initial vaccine series, partially vaccinated, and not vaccinated. Completion of the initial vaccine series was defined as patients who received the doses recommended by the CDC,15 depending on underlying illness and immune status. The not vaccinated population comprised patients who had never been vaccinated against COVID-19. Patients who did not belong to either group were classified as partially vaccinated. If patients had immunocompromised conditions, patients who received an additional dose (third dose in mRNA vaccine or ChAdOx1 nCoV-19, or second dose in Ad26.COV2-S) to complete primary series vaccination against COVID-19 were defined as completing the initial vaccine series according to CDC recommendation.15 If patients who completed the initial vaccine series received additional booster dose after final dose to complete vaccination, those patients were classified as first boosted vaccinated.
The first day of symptom onset was defined as “Day 0” if patients were initially symptomatic. Some patients were hospitalized due to concern of worsening, these patients were asymptomatic at the time of enrollment. In these cases, the date of diagnosis was considered as “Day 0” in asymptomatic patients. Information about the time of symptom onset was obtained via interview.
Variants of SARS-CoV-2 were identified with a genomic viral RNA Mini Kit (Qiagen Inc.), and classified by a double-multiplex RT-PCR assay using a PowerCheck SARS-CoV-2 S-gene mutation detection kit (Kogene Biotech Co., Ltd). Delta variants were defined as detection of both the P681R and L452R mutations, and Omicron variants as detection of the E484A, N501Y, and K417N mutations.
Detection of viable virus was defined as plaque formation in culture-based virus isolation. For the sake of logical validity, culture-negative samples or samples with copy numbers below the 95% limit of detection (LOD) (<2.6 copies/ml) collected before a culture- or PCR-positive sample were considered invalid and excluded from the survival analysis.
2.3 Sample collection and handling
All study patients were instructed to submit daily saliva samples and avoid toothbrushing, drinking, or eating before collecting saliva. Saliva samples were collected during their hospitalization, and we followed-up on all patients until their discharge. Collected saliva samples were stored at −80°C, and genomic-PCR and RT-PCR were performed. RT-PCR results are reported as Ct (cycle thresholds) and viral copy numbers. Viral copy numbers <2.6 log copies/ml were considered negative results, a 95% LOD. Culture-based virus isolation was performed on PCR-positive samples. Details of RT-PCR and culture-based virus isolation can be found in our previous reports.16, 17
2.4 Detection of genomic and subgenomic RNA by real-time RT-PCR assay
Subgenomic RNAs are discontinuous transcriptomes of coronaviruses which contain a nested set of negative-sense RNAs from the 3’ end of the virus genome joined to a common leader sequence derived from the 5’ end of the genomic RNA.18-20 Previous studies reported that the detection of subgenomic RNAs may better reflect viable virus than detection of genomic RNA.21, 22 Therefore, this study detected both of genomic and subgenomic RNA as well as culturable virus to understand association between transmission and viable virus of SARS-CoV-2 variants.
Viral RNA was extracted from respiratory specimens using a QIAamp viral RNA Mini kit (Qiagen Inc.) following manufacturer's instruction. To detect genomic RNA, PCR reaction mixture (20 μl) included 0.1 μl of 200× enzyme mix, 4 μl of 5× master mix (LightCycler Multiplex RNA Virus Master; Roche), 500 nM of each S and N gene primer, 200 nM of S and 250 nM of N gene probes, 250 nM of internal control primers, and 125 nM of internal control probes (Supporting Information: Table 1). To detect subgenomic RNA, the reaction mixture included 0.1 μl of 200× enzyme mix, 4 μl of 5× master mix, 1000 nm of leader primer, 500 nM of each S and N gene reverse primer, 250 nM of S and N gene probes, and internal control primers and probe (Supporting Information: Table 2). In each mixture, 5 μl of extracted RNA or in vitro-synthesized control RNA were added. PCR amplification was performed with a LightCycler 96 system (Roche) in the following conditions: reverse transcription at 50°C for 10 min, initial denaturation at 95°C for 5 min, 45 cycles of 2-step amplification, denaturation at 95°C for 10 s and annealing and elongation at 60°C for 30 s, and final extension at 60°C for 5 min. Calibration curves were generated using serial dilutions from 107 to 5 copies/μl of synthetic control RNA were assayed in six independent sets of reactions. The detection limit of this assay was 5 copies/reaction (2.6 log copies/ml of specimen) and viral copy numbers were determined by plotting the Ct values against log copies/reaction.12 The decision of positive and measurement of viral loads were determined by N gene of SARS-CoV-2. The sequences of primers and probe of genomic N gene were conserved 100% (66/66 bp) with omicron BA.1 and 98.5% (65/66 bp) with delta and omicron BA.2. Also, the sequences of primers and probe of subgenomic N gene were conserved 100% with all three variants (Supporting Information: Table 3, Figure 1).
2.5 Virus culture
Virus culture of SARS-CoV-2 from the saliva specimens with positive genomic RNA results was obtained using a plaque assay in a Biosafety Level 3 laboratory located at Korea University College of Medicine. Culture of Vero cells was done using six-well plates at a density of 9 × 105 cells/well for 24 h. Specimens were serially diluted 10-fold using PBS, and 200 μl of the diluted samples were inoculated into Vero cells and incubated for 1 h (37°C, 5% CO2) with rocking every 15 min, and overlaid with 2 ml of Dulbecco's Modified Eagle Medium/Nutrient Mixture F12 (DMEM/F-12) medium containing 0.6% oxoid agar. Viral plaque formation was visualized using crystal violet staining after 72 h of incubation at 37°C in a 5% CO2 incubator.
2.6 Secondary attack rates
Secondary attack rates were assessed by a study investigator (S. W. K.) within 1 week of diagnosis through face-to-face or wireline interviews based on records of epidemiologic investigation. We investigated the number of close contacts of each patient during their infectious period (≤2 days before symptom onset until the isolation in symptomatic patients or ≤2 days before the day of diagnosis until the isolation in asymptomatic patients), and the number of contact individuals who were diagnosed with SARS-CoV-2 infection within 1 week of contact. Close contacts were defined as individuals present within 6 feet of the confirmed case for at least 15 min. A more detailed definition of close contact has been given previously.23
2.7 Statistical analysis
All categorical variables were analyzed with χ2 and Fisher's exact tests, while continuous variables were processed by Wilcoxon sum tests. The Mantel−Haenszel test was used to adjust household and non-household secondary infections. To compare the viable virus shedding periods of Delta and Omicron variant, survival analysis and stratified Cox's proportional hazard models were constructed. For the multivariable analysis, variables with p < 0.10 in univariate analysis were selected. The Schoenfeld test was used to evaluate the constructed Cox proportional hazard model. Two-tailed p < 0.05 were considered statistically significant. All statistical analyses were performed with R for statistics (ver 4.2.4) and the Epi Info application.
3 RESULTS
3.1 Baseline characteristics and secondary attack rates
A total of 88 patients who agreed to submit daily saliva samples and were interviewed during the study period were enrolled. Of these patients, 6 were excluded from the analysis: the saliva specimens of 5 yielded undetectable or inconclusive RNA PCR results, and another was suspected of being coinfected with both the Delta and Omicron variants. Hence 82 patients were finally analyzed; 48 (59%) with Delta infections and 34 (41%) with Omicron infections. Baseline clinical characteristics of these subjects are shown in Table 1. There were no significant differences in terms of clinical characteristics between the two groups, except for vaccination status, treatments for COVID-19, duration of hospitalization, and duration from symptom onset to hospitalization.
| Characteristics | Delta (N = 48) | Omicron (N = 34) | p Value |
|---|---|---|---|
| Age, median years (IQR) | 60 (40−66) | 67 (52−75) | 0.054 |
| Male sex | 24 (50) | 23 (68) | 0.11 |
| Severity | 0.11 | ||
| Asymptomatic | 11 (23) | 17 (50) | |
| Mild | 17 (35) | 10 (29) | |
| Moderate | 7 (15) | 3 (9) | |
| Severe | 11 (23) | 3 (9) | |
| Critical | 2 (4) | 1 (3) | |
| Vaccination status | <0.001 | ||
| None | 27 (56) | 10 (29) | |
| Partially vaccinated | 4 (8) | 1 (3) | |
| Completion of the initial vaccine series | 16 (33) | 9 (26) | |
| First boostered | 1 (2) | 14 (41) | |
| Duration from the last vaccination, median days (IQR) | 79 (44−128) | 108 (73−164) | 0.40 |
| Duration from the symptom onset to hospitalization, median days (IQR) | 6 (3−11) | 2 (1−5) | <0.001 |
| Hospital duration, median days (IQR) | 6 (3−11) | 3 (3−5) | 0.001 |
| Underlying illness | |||
| Diabetes mellitus | 15 (32) | 12 (35) | 0.70 |
| Hypertension | 18 (38) | 13 (38) | >0.90 |
| Chronic kidney disease | 5 (11) | 7 (21) | 0.20 |
| Chronic liver disease | 3 (6) | 2 (6) | 0.90 |
| Cerebrovascular accident | 9 (19) | 6 (18) | 0.90 |
| Immunocompromised | 8 (17) | 4 (12) | 0.80 |
| Solid tumor | 12 (26) | 10 (29) | 0.70 |
| Treatment | |||
| Steroid | 22 (46) | 15 (44) | 0.70 |
| Immunomodulator | 18 (38)a | 4 (12)b | 0.008 |
| Remdesivirc | 24 (50) | 33 (97) | <0.001 |
| Monoclonal antibodyd | 25 (52) | 2 (6) | <0.001 |
| Secondary attack rates, no of infected individuals/no of contact (%) | 34/456 (7) | 38/124 (31) | <0.001 |
- Note: Data are presented as number of patients (%) unless otherwise indicated.
- a Thirteen patients received tocilizumab, whereas 4 patients received baricitinib. The remaining 1 patient received baricitinib and tocilizumab.
- b Two patients received tocilizumab, and the remaining 2 patients received baricitinib.
- c All patients received remdesivir as an antiviral agent. None received nirmatrevir/ritonavir or molnupiravir because these drugs were unavailable during the study period.
- d All patients received regdanvimab.
The Omicron patients had a higher secondary attack rate than the Delta patients (31% [38/124] vs. 7% [34/456], p < 0.001), and this difference persisted (p < 0.001 in Mantel−Haenszel test) after adjusting for vaccination status.
3.2 Viral kinetics
Figure 1 shows genomic and subgenomic viral copy numbers and virus culture positivity/negativity for individual patient samples (A,B, D,E), and daily percentages of culture positivity (C,F) for the Delta and Omicron infections, as functions of time from symptom onset. None of the samples from Delta patients were culture-positive beyond symptom onset Day 15, and none from Omicron patients were culture-positive beyond symptom onset Day 13.
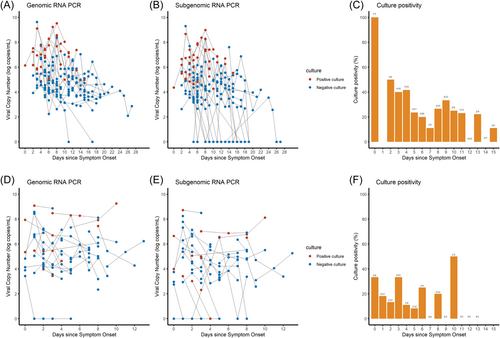
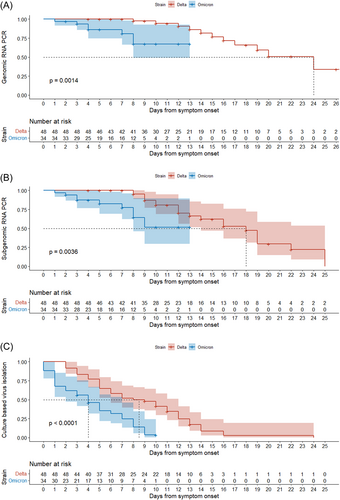
In the log-rank test, the Omicron variant had a shorter period of positivity for genomic RNA PCR and subgenomic RNA PCR than the Delta variant, while more than 50% of participants in Delta group did not reach negative conversion of genomic and subgenomic RNA PCR (median NA, IQR [8 to NA] vs. 24 days, IQR [16 to NA], p < 0.001: Figure 2A, median NA, IQR [8 to NA] vs. 18 days, IQR [12–22], p < 0.001: Figure 2B). The Omicron variant also had a shorter viable virus shedding period than the Delta variant (median 4, IQR [1–7] vs. 8.5 days, IQR [5–12 days], p < 0.0001, Figure 2C). Univariate and multivariable analyses of the factors associated with viable virus shedding are shown in Table 2. Moderate to critical disease severity (HR: 1.96, 95% CI: 1.06–3.57, p = 0.030) and immunocompromised status (HR: 2.17, 95% CI: 1.01–4.76, p = 0.046) were associated with longer viable virus shedding periods, whereas completion of the initial vaccine series or first boostered status (HR: 0.49, 95% CI: 0.28–0.86, p = 0.015) and Omicron infection (HR: 0.44, 95% CI: 0.23–0.81, p = 0.009) were associated with shorter viable virus shedding periods in multivariable analysis. Adjusted Cox proportional hazard survival curves for viable virus shedding after adjustment of confounders in Omicron and Delta infections are shown in Supporting Information: Figures 2 and 3. Survival curves adjusted simultaneously for disease severity, vaccination status and immune status confirmed the shorter viable virus shedding period of the Omicron variant (p = 0.02). The shorter viable virus shedding period of the Omicron variant also persisted after separate adjustment for each of these confounders: vaccination status (p = 0.002), immune status (p < 0.001), and disease severity (p < 0.001). Furthermore, since COVID-19 treatments were highly correlated with disease severity, we performed additional analysis including COVID-19 treatment in the final model (Supporting Information: Tables 4 and 5). These two models consistently revealed that Omicron infection was significantly associated with shorter viable viral shedding.
| Characteristics | N | Univariate analysis | Multivariable analysis | ||||
|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% confidence interval | p Value | Adjusted hazard ratio | 95% confidence interval | p Value | ||
| Age | 82 | 1.00 | 0.98–1.00 | 0.50 | |||
| Male sex | 47 | 1.02 | 0.63–1.63 | >0.90 | |||
| Severity | |||||||
| Asymptomatic to mild | 55 | Reference | Reference | ||||
| Moderate to critical | 27 | 2.04 | 1.19–3.48 | 0.009 | 1.96 | 1.06–3.57 | 0.030 |
| Vaccination status | |||||||
| Not or partially vaccinated | 42 | Reference | - | Reference | - | ||
| Completion of the initial vaccine series or first boosted | 25 | 0.39 | 0.24–0.63 | <0.001 | 0.49 | 0.28–0.87 | 0.015 |
| Duration from the last vaccination | 42 | 1.00 | 0.99–1.00 | 0.20 | |||
| Variant | |||||||
| Delta | 47 | Reference | - | Reference | - | ||
| Omicron | 34 | 0.32 | 0.19–0.54 | <0.001 | 0.44 | 0.23–0.81 | 0.009 |
| Underlying illness | |||||||
| Diabetes mellitus | 24 | 0.94 | 0.57–1.56 | 0.80 | |||
| Hypertension | 29 | 1.07 | 0.66–1.72 | 0.80 | |||
| Chronic kidney disease | 10 | 0.81 | 0.43–1.51 | 0.50 | |||
| Chronic liver disease | 4 | 0.67 | 0.24–1.89 | 0.40 | |||
| Cardiovascular accident | 13 | 1.01 | 0.54–1.89 | >0.90 | |||
| Immunocompromised | 15 | 2.04 | 0.99–4.16 | 0.053 | 2.17 | 1.01–4.76 | 0.046 |
| Solid tumor | 26 | 0.57 | 0.33–0.97 | 0.039 | |||
4 DISCUSSION
In this study, we have compared the secondary attack rates and viral kinetics of the Delta and Omicron variant in hospitalized patients with various degrees of severity of COVID-19 infection. We found that Omicron infection was associated with a higher secondary attack rate, despite the fact that the viable virus shedding period was only half that of the Delta variant. Omicron infection was also independently associated with a shorter viable virus shedding period after adjustment of disease severity, immunocompromised condition, and vaccination status. Our findings provide an important observational basis for the choice of isolation period for patients with Omicron infections, as well as indications of what biologic factors may contribute to the higher transmissibility of this variant.
Previous studies comparing the viable virus shedding periods of the Omicron and Delta variants found no significant difference between them (median 5 days in the Delta group vs. 4 days in the Omicron group,5 median 3 days in the Delta group vs. 2 days in the Omicron group6) which is not consistent with our present results (Figure 2 and Table 2). The shorter viable virus shedding period of patients with the Delta variant (median 5−6 days) is in line with our previous results in healthy young adults with SARS-CoV-2 Delta infections.16, 17 However, a few of the patients in those studies were symptomatic and most had completed the initial vaccine series, and since disease severity and vaccination status are correlated with duration of viable virus shedding,14, 24, 25 the low proportion of symptomatic patients in those study populations, and the absence of adjustment of vaccination status, may have led to the failure to detect the difference of viable virus shedding period between Delta and Omicron variants. We should point out that our finding supports the CDC's recommendation to reduce the isolation period for Omicron infections to 5 days after symptom onset apart for Delta infections,26 since our data clearly demonstrated that viable viral shedding period in Omicron infection was shorter than in the Delta infection. The reasons for this different viral shedding kinetics are not clear. Omicron variants achieve immune evasion effects via spike protein mutation; on the other hand, it compromises cell entry in TMPRSS-2 expressing cells and ability to form syncytia.27 In addition, previous studies suggested that syncytia may facilitate the viral spread and contribute in modifying the disease severity,28 therefore impaired syncytia formation with Omicron infection may lead to shortened viral shedding period in Omicron variants. Further studies are needed in this area.
The high secondary attack rates observed for the Omicron variant are consistent with the results of other studies.2-4 However, there is an unsettled question about which biological factors contribute to this high transmissibility. Our data indicate that it does not stem from prolonged viable viral shedding. Possible explanations are (1) low infectious dose, (2) faster replication in the human respiratory epithelium, (3) more exhalation as tiny aerosols, (4) increased stability in the air, and (5) high transmissibility in the asymptomatic or presymptomatic periods.29-31 The Omicron variant harbors multiple spike protein mutations that increase its affinity for the ACE2 receptor and make it less dependent on the host protease.27 Those changes contribute to its low infectious dose. It also has increased environmental stability, suggesting that transmission via the airborne route may be more frequent.32, 33 It may also replicate faster in human respiratory epithelial cells,34 and this could lead to more rapid accumulation of mutations and intrahost evolution in chronic infections.35, 36 Recently, Lai et al. found that viral RNA shedding of exhaled breath aerosols in patients with Alpha, Delta, and Omicron variants increased, compared with previous variants of concerns, but there was no significant difference in peak RNA shedding between Delta and Omicron variants.36 Interestingly, the highest shedder had an Omicron infection and shed viral RNA copies three times more than the maximum level observed for Delta and Alpha infections.36 This finding suggests that the Omicron variant is more likely to produce super-spreader. Finally, its patent period may begin before symptom onset: Ke et al.7 have proposed that transmission of SARS-CoV-2 occurs predominantly during the presymptomatic phase. Further studies of this important issue are needed to prepare for the emergence of further highly transmissible variants.
This study has several limitations. First, since it involved only patients admitted to a tertiary hospital, it is likely to have included more severe disease; this makes it hard to generalize our findings to individuals with asymptomatic SARS-CoV-2 infections or mild COVID-19. It is worth to note that there was a significant difference in duration from symptom onset to hospitalization between Delta and Omicron groups (Table 1). The difference of severity of COVID-19 between two groups may partially explain this phenomenon. The multivariate analysis with the adjustment of the disease severity revealed that patients with Omicron infections had significantly shorter duration of viable viral shedding. Despite this, there might be unmeasured confounders which affected the hospitalization delay during Delta-dominant period. Second, despite the request for all participating patients to provide daily saliva samples, some were lost to nonadherence, causing interval censoring. Fortunately, this applied to only three Delta samples (two for 1 day and one for 2 days) and three Omicron samples (two for 1 day and one for 5 days) all in cases of negative conversion. Therefore, interval censoring is not considered to have significantly affected our main findings. Third, we used saliva samples for viable viral shedding kinetics instead of nasopharyngeal (NP) swab samples because saliva specimen collection had the advantage of dense sampling. Variability in Ct values, even within the same day was shown with saliva specimens.37 In addition, the SARS-CoV-2 viral load was lower in saliva samples than in NP swab samples.38 However, a meta-analysis revealed that the sensitivity of the two types of samples was comparable.39 Our previous studies also demonstrated that daily saliva specimens might be the alternative for our understanding of viable viral kinetics.16, 17 Fourth, some may argue that prior asymptomatic infection may contribute to virus shedding kinetics. Although we excluded patients with prior history of COVID-19, we did not perform the N protein antibody test, so prior subclinical or asymptomatic SARS-CoV-2 infection could in principle have had a confounding effect. However, since for most of the study period the prevalence of COVID-19 in South Korea remained below 1% of the total populations,40 it is unlikely to have had any major effect. Finally, although none of the participants received nirmatrelvir-ritonavir in this study, recent studies reported that some patients with nirmatrelvir-ritonavir experienced rebound phenomenon, and there was evidence of high viral load and viable viral shedding during this rebound period.41, 42 Therefore, further studies are needed to understand the effect of nirmatrelvir-ritonavir on viable viral shedding in patients with Omicron infection.
In conclusion, patients with Omicron infections had a higher transmission rate but a shorter period of shedding of transmissible virus than patients with Delta infections. Our findings support the current CDC recommendation of a 5-day isolation period for Omicron patients. Further studies are needed of the basis of the high transmissibility of the Omicron variant during the early symptomatic period.
AUTHOR CONTRIBUTIONS
Sung-Woon Kang, Ji Yeun Kim, and Heedo Park contributed equally. The study was designed by Sung-Han Kim and Man-Seong Park. This study was supervised by Euijin Chang, Seongman Bae, Jiwon Jung, Jeonghun Kim, Yong Pil Chong, Sang-Oh Lee, Sang-Ho Choi, and Yang Soo Kim. The manuscript was written by Sung-Woon Kang and Ji Yeun Kim. The data collection, analysis, and interpretation were performed by Sung-Woon Kang, Ji Yeun Kim, Heedo Park, So Yun Lim, and Jeonghun Kim. All authors were involved in the critical revision of the manuscript.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the contribution of the healthcare workers of Asan Medical Center. This study was supported by grants from the research fund donation for COVID-19 research to Asan Medical Center by Kyu-Kang Cho, the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), which is funded by the Ministry of Health & Welfare, Republic of Korea (grant No. HD22C2045), and from the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT, Republic of Korea (NRF-2022M3A9I2017241).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data in present study are available from the corresponding author upon reasonable request.




