Establishment of an indirect ELISA method for antibody detection of porcine pseudorabies by recombinant gB, gC, and gD proteins
Abstract
Pseudorabies virus (PRV), as a neuroherpes virus, leads to heavy economic losses in the pig industry worldwide. This study was designed to establish recombinant PRV glycoprotein B (gB), C, and D proteins as PRV diagnostic antigens. The gB/C, gC/D, and gB/C/D fusion sequences were synthesized and inserted into pET-28a+ vector to generate the recombinant plasmids. The identified positive recombinant plasmids were transformed into BL21 Escherichia coli. The results of the polymerase chain reaction and enzyme digestion showed that the gB/C, gC/D, and gB/C/D fusion proteins were successfully expressed. An indirect sandwich ELISA was developed with the gB/C, gC/D, and gB/C/D as coating antigens. The results of indirect enzyme-linked immunosorbent assay (ELISA) analysis of 184 PRV-positive porcine sera showed that the positive coincidence rates of three recombinant proteins ELISAs relative to IDEXX kit were 98.25%, 95.32%, and 98.83%, and the negative coincidence rates were 85.71%, 75% and 100%, respectively. The inter and intra batch repeatability tests showed that the coefficient of variations of our kits were all less than 5%. Especially, the gB/C/D-ELISA has the highest specificity and sensitivity among the ELISA methods developed in this study. We established a series expression system of gB/C, gC/D, and gB/C/D antigen epitope genes and Recombinant protein-based indirect ELISA, providing new ideas for PV diagnosis and vaccine development.
1 INTRODUCTION
Pseudorabies virus (PRV), as a neuroherpes virus, can infect livestock and wild animals such as pigs, cattle, and sheep. It reduces livestock productivity and even causes death by invading the nervous and reproductive systems.1, 2 Since 2011, with the emergence of PRV mutants, PR has broken out in many large pig farms in China, resulting in sow abortion, piglet neurological symptoms, and high mortality.1 At present, the prevalence of PR caused serious economic losses to the pig industry of China.3-5
PRV glycoproteins not only play an important role in the process of virus invasion, release and intercellular diffusion, but also have also been identified as the main target proteins involved in the host immune response.6, 7 The gB, gC, and gD glycoproteins are important envelope components of mature virus particles among the 11 envelope glycoproteins encoded by PRV.6 They participate in the process of virus adsorption and penetration, and serve as the most important protective antigens to stimulate the host to produce neutralizing antibodies and initiate virus-specific cellular immune responses.6 Studies have shown that subunit vaccines based on gB, gC, and gD proteins can protect mice or pigs from PRV infection.8-10 The three are the preferred proteins for developing vaccines immunization and antibody detection kits with high immunogenicity and reactivity.11-13
In the present research, gB, gC, and gD were fused and expressed in different combinations. On this basis, we established new PRV neutralizing antibody detection methods, providing an experimental basis for PV diagnosis and vaccine development.
2 MATERIALS AND METHODS
2.1 Reagents
The reagents used in this study, 2×EasyTaq® PCR SuperMix (+dye) (AS111), 2×TransStart® FastPfu Fly PCR SuperMix (-dye) (AS231), Trans DNA Marker II (BM101), Trans8K DNA Marker (BM151), FlyCut® EcoRI (JE201), FlyCut® NcoI (JN101), EasyPure® Quick Gel Extraction Kit (EG101), T4 DNA Ligase (FL101), EasyPure® Plasmid MiniPrep Kit (EM101), Agarose, GelStain, BL21 (DE3) Chemically Competent Cell (CD601) and ProteinRuler® II (12–120 kDa) (DR201) were all purchased from TransGen Biotech.
2.2 Sequences and plasmids
The fusion genes were designed based on the sequences of PRV gB, gC, and gD genes. The gB aa 59–126, gC aa 156–290, and gD aa 78–183 were selected for reorganization. According to the properties of amino acids, 2–4 glycines were inserted between the epitopes, and then the epitopes were connected in series on the ORF without signal peptide. Sequences of PRV-gB/C, gC/D, and gB/C/D were all synthesized by Biosune Biotechnology Co., Ltd. The recombinant plasmids were validated using sequencing in Sangon Biotech. The synthesized sequences were inserted into the pET-28a+ vector (between NcoI and EcoRI) to construct the recombinant plasmids.
2.3 Polymerase chain reaction (PCR) and enzyme digestion identification
PCR was preformed following the instructions. Details of the PCR kit were in the reagents section. The primers used in this study were synthesized from Biosune Biotechnology Co., Ltd., including gB/C (F-CCCAATCCATGGCCGCAGCAGTTACCCGTGCAG and R-CCCAATGAATTCGGAAACGCGGCATCAACCGGA), gC/D(F-CCCAATCCATGGCCTATTTTGATGAACCGCCGCGT and R-CCCAATGAATTCGGCAGGGCAACCATAAAATCTGTCAG) and gB/C/D (F-GCAGCAGTTACCCGTGCAGCAA and R-CAGGGCAACCATAAAATCTGTCAG).
The extracted recombinant plasmids were verified by double enzyme digestion according to the instructions. After added 1 μl/50 μl NcoI and EcoRI, plasmids were incubated at 37°C for 15 min. Double-enzyme digestion products were detected by agarose gel electrophoresis of 1.5%.
2.4 Induction of protein expression
Three verified strains were seeded in 5 ml LB/Kan+ medium and cultured at 37°C with 250 rpm. When OD600 reached 0.5, 1 mM IPTG was added to induce protein expression. After the induction for 2, 4, and 6 h, the bacterial solution was collected and then boiled in a protein loading buffer at 95°C for 5–10 min.
2.5 SDS-PAGE and western blot
The protein was separated with 5% concentrated gel and 12% separated gel. Thirty micrograms of protein samples were loaded to the lane and electrophoresed at 70 V for 30 min and then 90 V until the indicator reached at about 0.5 cm from the lower end of the gel. The samples were stained with Coomassie brilliant blue at room temperature for 1 h. After decolorization, photos were taken.
For western blot analysis, the proteins were transferred from sodium dodecyl sulfate polyAcrylamide gel electrophoresis to polyvinylidene fluoride (PVDF) membranes at 200 mA for 90 min. After blocking with 5% bachelor of science in agriculture at room temperature for 2 h, PVDF membranes were incubated with primary antibodies at 4°C overnight, followed by incubation with secondary antibodies at room temperature for 2 h. The proteins were maintained with chemiluminescence reagent for 2 min and then were exposed for photography. Details of antibodies were shown in the reagents section.
2.6 Establishment of indirect ELISA for recombinant protein
According to matrix titration, different concentrations of recombinant proteins (0.5, 1, 3, and 5 mg/L) and serums with different dilutions (1:20, 1:50, 1:100, 1:200) were added to 96-well plates and incubated at 37°C for 60 min. Recombinant proteins were incubated with Enzyme labeled antibody at 37°C for 60 min. Then, 100 μl of TMB substrate chromogenic solution was added and maintained at 37°C for 10 min. OD650 value was measured by a microplate reader.
3 RESULTS
3.1 Construction of recombinant expression vectors
First, the gB/C, gC/D, and gB/C/D fusion sequences were designed and synthesized according to the sequence information in NCBI. Then, the target sequences were cut off and connected to the prokaryotic expression vector pET-32a. The target bands were successfully amplified by PCR assay (Figure 1A).
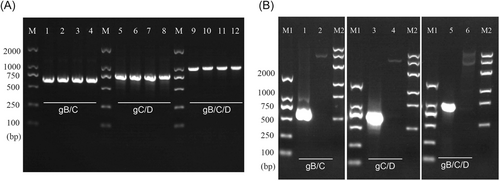
The accuracy of the synthesized sequence was identified by enzyme digestion and sequencing. As shown in Figure 1B, after double enzyme digestion, the fragment lengths of gB/C, gC/D, and gB/C/D were consistent with the expected result. According to the sequencing results shown in Supporting Information: Figure, the bases of the BC and CD fragments were completely consistent with those of wild-type fragments, and the bases of the gB/C/D fragment and the wild-type fragment are more than 99% identical.
3.2 Expression of recombinant proteins
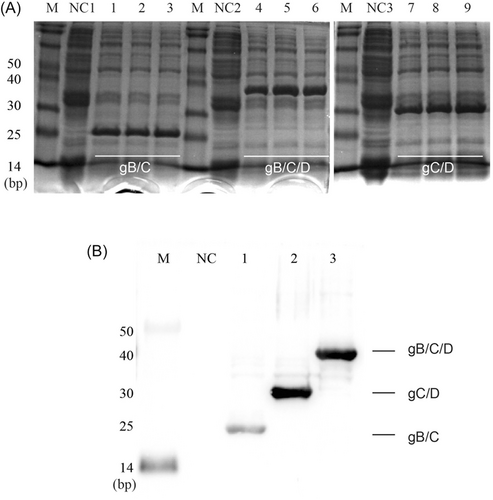
The identified recombinant plasmids were transformed into BL21 Escherichia coli competent cells to express the recombinant proteins. IPTG was used to induce protein expression for 0, 2, 4, and 6 h. According to the software calculation, the molecular weights of gB/C, gC/D, and gB/C/D recombinant proteins were 21.51, 27.17, and 33.92 kDa, respectively. As shown in Figure 2A, the molecular weight of recombinant proteins gB/C, gC/D, and gB/C/D were all consistent with the predicted molecular weight. Then, the purified protein was analyzed by western blot. The results showed that specific bands were displayed, indicating that BL21 strain successfully expressed recombinant proteins (Figure 2B).
3.3 Establishment of indirect ELISA based on recombinant proteins
Then, we constructed an indirect ELISA method based on these recombinant proteins for the detection of porcine pseudorabies (Figure 3). The purified diluted fusion proteins (0.5, 1, 3, 5 mg/L) were coated as antigen, and the negative and positive sera were diluted at the ratio of 1:20, 1:50, 1:100, and 1:200 to optimize conditions. As shown in Table 1, the concentrations of fusion proteins gB/C and gC/D were 3 mg/L, and the sample serums were diluted at 1:50 to maximize the P/N value; the concentration of fusion protein gB/C/D was 1 mg/L, and the sample serum was diluted at 1:50 to maximize the P/N value.
| Protein (mg/L) | 1:20 | 1:50 | 1:100 | 1:200 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 3 | 1 | 0.5 | 5 | 3 | 1 | 0.5 | 5 | 3 | 1 | 0.5 | 5 | 3 | 1 | 0.5 | |
| gB/C | ||||||||||||||||
| P | 0.902 | 0.835 | 0.780 | 0.712 | 0.883 | 0.821 | 0.759 | 0.664 | 0.833 | 0.780 | 0.672 | 0.611 | 0.799 | 0.746 | 0.658 | 0.607 |
| N | 0.413 | 0.396 | 0.379 | 0.350 | 0.400 | 0.360 | 0.348 | 0.324 | 0.389 | 0.349 | 0.333 | 0.304 | 0.379 | 0.353 | 0.323 | 0.305 |
| P/N | 2.184 | 2.109 | 2.058 | 2.034 | 2.208 | 2.281 | 2.181 | 2.049 | 2.141 | 2.235 | 2.018 | 2.010 | 2.108 | 2.113 | 2.037 | 1.990 |
| gC/D | ||||||||||||||||
| P | 0.891 | 0.829 | 0.761 | 0.699 | 0.863 | 0.801 | 0.715 | 0.633 | 0.811 | 0.750 | 0.643 | 0.592 | 0.780 | 0.723 | 0.646 | 0.599 |
| N | 0.409 | 0.388 | 0.361 | 0.336 | 0.397 | 0.359 | 0.339 | 0.306 | 0.379 | 0.341 | 0.320 | 0.295 | 0.365 | 0.348 | 0.326 | 0.303 |
| P/N | 2.178 | 2.137 | 2.108 | 2.080 | 2.174 | 2.231 | 2.109 | 2.069 | 2.140 | 2.199 | 2.009 | 2.007 | 2.137 | 2.078 | 1.982 | 1.977 |
| gB/C/D | ||||||||||||||||
| P | 0.946 | 0.889 | 0.847 | 0.762 | 0.903 | 0.843 | 0.811 | 0.708 | 0.885 | 0.822 | 0.743 | 0.699 | 0.818 | 0.765 | 0.683 | 0.626 |
| N | 0.423 | 0.412 | 0.375 | 0.356 | 0.415 | 0.400 | 0.350 | 0.331 | 0.399 | 0.365 | 0.336 | 0.323 | 0.387 | 0.359 | 0.325 | 0.301 |
| P/N | 2.236 | 2.158 | 2.259 | 2.140 | 2.176 | 2.108 | 2.317 | 2.139 | 2.218 | 2.252 | 2.211 | 2.164 | 2.114 | 2.131 | 2.102 | 2.080 |
3.4 Validity verification for indirect ELISA based on recombinant proteins
184 cases of PVR-positive pig sera were collected to verify the specificity of the indirect ELISA method obtained in the above experiment. As shown in Table 2, of the 184 samples, 171 were diagnosed as positive and 6 cases were misjudged as negative by IDEXX kit; 169 cases were diagnosed as positive and 6 cases were misjudged as negative by gB/C/D-ELISA; 163 cases were diagnosed as positive and 8 cases were misjudged as negative by gC/D-ELISA; 168 cases were diagnosed as positive and 7 cases were misjudged as negative by gB/C-ELISA. The positive coincidence rates of three recombinant proteins ELISA and IDEXX kit were 98.25%, 95.32%, and 98.83%, respectively; the negative coincidence rates were 85.71%, 75%, and 100%, respectively. Statistical analysis showed that there was no significant difference in the positive rate between the three fusion proteins and IDEXX kit. Importantly, gB/C/D-ELISA had the highest specificity and sensitivity among the fusion protein ELISAs developed in this study, but it still needed to be improved.
| Method | Samples (n) | Positive rate | Negative rate | Suspicious rate | Coincidence rate | ||
|---|---|---|---|---|---|---|---|
| PRV+ | PRV− | Suspicious | |||||
| gB/C-ELISA | 168 | 7 | 9 | 91.30% | 3.80% | 4.89% | 98.37% |
| gC/D-ELISA | 163 | 8 | 13 | 88.59% | 4.35% | 7.07% | 95.65% |
| gB/C/E-ELISA | 169 | 6 | 9 | 91.85% | 3.26% | 4.35% | 98.91% |
| PRV-gB Kit (IDEXX) | 171 | 6 | 7 | 92.93% | 3.26% | 3.80% | / |
- Note: Coincidence rate = (positive + negative + suspicious)/total sample size × 100%.
- Abbreviations: ELISA, enzyme-linked immunosorbent assay; PRV, pseudorabies virus.
To clarify the repeatability of the ELISA established in this study, intra batch and inter batch repeatability tests were performed on five randomly selected serum samples.14 For gB/C-ELISA, the intra batch coefficient of variation (CV) was 1.8%, and the inter batch CV was 4.3%, which were less than 5%, indicating that the gB/C-ELISA method was reproducible (Table 3). As shown in Table 4, the intra batch CV was 1.8%, and the inter batch CV was 3.4%, suggesting good repeatability of gC/D-ELISA. For gB/C/D-ELISA, the intra batch CV was 1.8%, and the inter batch CV was 3.0%, which also were less than 5% and indicated a high repeatability of gB/C/D-ELISA (Table 5). Therefore, the ELISA methods established in this study had good repeatability.
| Sample | OD650 | OD650 | OD650 | Mean | SD | CV | CV mean |
|---|---|---|---|---|---|---|---|
| Intra batch | |||||||
| 1 | 0.668 | 0.661 | 0.676 | 0.008 | 0.668 | 1.1% | 1.8% |
| 2 | 0.638 | 0.613 | 0.641 | 0.015 | 0.631 | 2.4% | |
| 3 | 0.73 | 0.759 | 0.736 | 0.015 | 0.742 | 2.1% | |
| 4 | 0.297 | 0.299 | 0.303 | 0.003 | 0.300 | 1.0% | |
| 5 | 0.288 | 0.301 | 0.295 | 0.007 | 0.295 | 2.2% | |
| Inter batch | |||||||
| 1 | 0.671 | 0.633 | 0.682 | 0.026 | 0.662 | 3.9% | 4.3% |
| 2 | 0.665 | 0.634 | 0.612 | 0.027 | 0.637 | 4.2% | |
| 3 | 0.761 | 0.733 | 0.706 | 0.028 | 0.733 | 3.8% | |
| 4 | 0.312 | 0.306 | 0.335 | 0.015 | 0.318 | 4.8% | |
| 5 | 0.293 | 0.297 | 0.319 | 0.014 | 0.303 | 4.6% |
- Abbreviations: CV, coefficient of variation; ELISA, enzyme-linked immunosorbent assay.
| Sample | OD650 | OD650 | OD650 | Mean | SD | CV | CV mean |
|---|---|---|---|---|---|---|---|
| Intra batch | |||||||
| 1 | 0.653 | 0.644 | 0.663 | 0.010 | 0.653 | 1.5% | 1.8% |
| 2 | 0.616 | 0.607 | 0.633 | 0.013 | 0.619 | 2.1% | |
| 3 | 0.736 | 0.743 | 0.752 | 0.008 | 0.744 | 1.1% | |
| 4 | 0.279 | 0.286 | 0.293 | 0.007 | 0.286 | 2.4% | |
| 5 | 0.283 | 0.295 | 0.291 | 0.006 | 0.290 | 2.1% | |
| Inter batch | |||||||
| 1 | 0.66 | 0.627 | 0.679 | 0.026 | 0.655 | 4.0% | 3.4% |
| 2 | 0.658 | 0.616 | 0.619 | 0.023 | 0.631 | 3.7% | |
| 3 | 0.739 | 0.748 | 0.703 | 0.024 | 0.730 | 3.3% | |
| 4 | 0.307 | 0.31 | 0.325 | 0.010 | 0.314 | 3.1% | |
| 5 | 0.279 | 0.293 | 0.295 | 0.009 | 0.289 | 3.0% |
- Abbreviations: CV, coefficient of variation; ELISA, enzyme-linked immunosorbent assay.
| Sample | OD650 | OD650 | OD650 | Mean | SD | CV | CV mean |
|---|---|---|---|---|---|---|---|
| Intra batch | |||||||
| 1 | 0.701 | 0.679 | 0.683 | 0.012 | 0.688 | 1.7% | 1.8% |
| 2 | 0.655 | 0.637 | 0.648 | 0.009 | 0.647 | 1.4% | |
| 3 | 0.737 | 0.766 | 0.743 | 0.015 | 0.749 | 2.0% | |
| 4 | 0.294 | 0.293 | 0.3 | 0.004 | 0.296 | 1.3% | |
| 5 | 0.285 | 0.297 | 0.298 | 0.007 | 0.293 | 2.5% | |
| Inter batch | |||||||
| 1 | 0.686 | 0.654 | 0.689 | 0.019 | 0.676 | 2.9% | 3.0% |
| 2 | 0.657 | 0.621 | 0.667 | 0.024 | 0.648 | 3.7% | |
| 3 | 0.757 | 0.725 | 0.736 | 0.016 | 0.739 | 2.2% | |
| 4 | 0.305 | 0.286 | 0.302 | 0.010 | 0.298 | 3.4% | |
| 5 | 0.296 | 0.282 | 0.283 | 0.008 | 0.287 | 2.7% |
- Abbreviations: CV, coefficient of variation; ELISA, enzyme-linked immunosorbent assay.
4 DISCUSSION
PRV contains 11 envelope glycoproteins, among which gB, gC, and gD glycoproteins are important envelope components of mature virions and involve in the process of virus adsorption and invading host cells. The gB, gC, and gD are the main protective antigens, that stimulate the host to produce neutralizing antibodies and induce virus-specific immune responses. The antibodies they stimulated were able to neutralize PRV in vivo and in vitro. In this study, the gB, gC, and gD genes of the mutant PRV strain FJ-2012 were used to realize the tandem expression of the antigenic epitopes of gB/C, gC/D, and gB/C/D glycoproteins. These recombinant proteins were used as coating antigen to detect PRV neutralizing antibody in pig serum by indirect ELISA.
First, the gB/C, gC/D, and gB/C/D fusion plasmids were constructed and successfully expressed in BL21. We selected the dominant epitope with high immunoreactivity of gB, gC, and gD as the target sequences. The epitope aa 59–126, which can specifically bind to mAbs, was selected for gB protein.15, 16 There are three consecutive epitope regions in aa 59–126 at the N-terminal of PRV gB glycoprotein with the characteristics of high flexibility and hydrophilicity.16 As the main Neutralizing Antigen, gC protein is one of the critical T cell antigens of PRV, and aa 156–290 of gC-M is the main antigen region. gC glycoprotein is necessary to mediate the adhesion between the virus and target cells. The three heparin binding regions located in the antibody binding region (aa 44–290) at the N end of gC glycoprotein. The binding of gC-specific antibody to this region can block the adhesion of the virus. The aa 78–183 of gD-N was selected in the present research. This regain is the critical antigen region of gD, which is also the main protective antigen of PRV.17 Therefore, the gB/C/D recombinant protein constructed in this study has the potential to be used as both immune antigen and detection method.
At present, the diagnostic strategies of PRV mainly include virus isolation, PCR, and serum antibody detection.18-21 However, virus isolation and culture generally take 2–3 days with poor sensitivity, which is mainly due to the inappropriate observation of attenuated strains and the difficulty in isolating cells with low virus content. Although PCR is a fast, accurate, and sensitive detection method, it is not suitable for clinical practice due to its high requirements for professional skills and expensive equipment. Compared with the above two methods, serum antibody detection is more suitable for batch detection and clinical practice with the advantages of simple operation, good sensitivity.22, 23 Indirect ELISA method based on the specific antibody is an effective strategy.24
Then, the PRV indirect ELISA antibody detection kits based on gB/C, gC/D and gB/C/D recombinant proteins were developed. One hundred and eighty-four cases of PVR-positive pig sera were collected to verify the specificity of these three ELISA methods. IDEXX gB Kit was used as the control kit.25 The positive coincidence rates of three recombinant proteins ELISA and IDEXX kit were 98.25%, 95.32%, and 98.83%, respectively; the negative coincidence rates were 85.71%, 75%, and 100%, respectively. In addition, the inter and intra batch repeatability tests showed that the CVs of our kits were all less than 5%. These results proved that the serological diagnostic method based on gB/C, gC/D, and gB/C/D fusion proteins could basically meet the requirements of differential diagnosis of PRV. Especially, gB/C/D-ELISA had the highest specificity and sensitivity among the fusion protein ELISA developed in this study.
Epitopes stimulate the host to produce antibodies or sensitized lymphocytes and recognize them, playing an important role in the structure and function of protein antigens.26 Tandem epitopes not only purify the antigenicity but also reduce the influence of irrelevant interference sequences. Tandem epitopes have been widely used in the preparation of diagnostic antigens and immunogens.27 In recent years, recombinant protein-based ELISA with multiple epitopes is developed as the virus detection vaccine candidate in Foot-and-mouth disease, bovine leukemia virus infection, and Duck Plague.1, 28-31 However, the clinical serum samples in this study are limited by regionalization. We will perform detection and comparative analysis in larger sample size, and investigate the immune protection effect of the purified recombinant protein as an immunogen in mice.
In conclusion, we established a series expression system of gB/C, gC/D, and gB/C/D antigen epitope genes, optimized accurate and sensitive PRV neutralizing antibody detection methods, and provided a new research idea for the development of efficient and safe PRV vaccine.
AUTHOR CONTRIBUTIONS
Xuemin Wu conceptualized and designed the study. All authors performed the experiments and analyzed the data. Xuemin Wu wrote the paper. Lunjiang Zhou and Longbai Wang responsible for the management and coordination of the planning and execution of research activities.
ACKNOWLEDGMENTS
This study was supported by Fujian Natural Science Foundation (2020J01350, 2020J01347, and 2019J02014), special program of Fujian public welfare scientific research institutes (2022R1026002).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




