Prevalence of drug resistant HIV-1 forms in patients without any history of antiretroviral therapy in the Republic of Guinea
Abstract
To study the structure of human immunodeficiency virus (HIV)-1 drug resistance (DR) in patients with newly diagnosed infection. Residents of the Republic of Guinea (N = 2168) were tested for HIV using enzyme-linked immunosorbent assay (ELISA). Individuals with a positive result were further examined for the presence of viral load in blood plasma. HIV was analyzed using Sanger sequencing. The obtained sequences were genotyped using REGA (version 3.0) and analyzed in MEGA 7. Analysis for the presence of DR mutations was performed using the Stanford University HIV DR Database. Serological markers of HIV were detected in 239 people, which represents 11.02% of the entire sample. HIV RNA was detected in 58 people. The following subtypes were seen: HIV CRF02_AG (41.9%); A1 (29.1%); A3 (12.9%); URF A1_G (12.9%); and G (3.2%). In 25% of patients, at least one significant mutation was encountered leading directly to HIV DR. The mutations encountered cause resistance to NRTI and NNRTI; one case of multiple resistance was identified. Major resistance to protease inhibitor was not seen. The detection of HIV-1 mutations associated with DR, in individuals who have never received antiretroviral therapy, is a cause for concern. It suggests that: new infections are occurring with strains that already have resistance; and the expansion of resistance is not always directly associated with selective drug pressure. Among the likely reasons for the high prevalence of primary HIV DR in the Republic of Guinea, drug availability is probably the key. The consequence of this is the lack of adherence of patients to treatment, the formation and transmission of resistant variants of the virus in the population. These findings suggest the need to test patients for resistant virus variants before initiating treatment.
1 INTRODUCTION
The human immunodeficiency virus (HIV) epidemic continues to spread rapidly around the world. Uncontrolled illness caused by HIV infection is a long-term, progressive infectious disease characterized by the presence of severe immunodeficiency, accompanied by a severe course of concomitant and opportunistic diseases.1, 2 According to Joint United Nations Program on HIV/AIDS (UNAIDS) estimates, the number of people currently infected with HIV globally is about 31.6−44.5 million, while the number of new infections amounted to 1.2−2.2 million cases in 2019.3 The African continent is one of the most HIV-affected regions in the world. It is currently home to 25.6 million people living with HIV (PLHIV), which is 67.37% of all registered HIV infections globally. The overwhelming majority (80.86%) are in the countries of Eastern and Southern Africa.3
The combination of mutational and recombinational processes in the evolution of HIV has led to the existence of the HIV diversity that is currently observed. There are three HIV-1 groups (M, N, O). Of these, group M includes all of the most common viral variants, while the other two represent only a small number of strains.4, 5 In group M, from 14 to 18 subtypes are distinguished based on whole genome analysis. These are denoted by the letters A, B, C, D, etc. The subtypes differ from each other by an average of 25%–30% by genomic nucleotide sequence.
In terms of the number of HIV infections, subtype C (prevalent in southern Africa and Asia) is the globally predominant subtype (47.2%). All HIV-1 subtypes were formed in Africa, with the exception of subtype B (whose final evolution ended outside the continent). Their subsequent uneven distribution over the globe is considered a consequence of migratory processes in the human population.4, 6
Currently, HAART is a combination of three, or less often four, antiretroviral drugs, the pathways of which are directed at different stages of HIV replication in the human body.7 When using HAART, doctors often face the problem of treatment failure. The most common reason for the ineffectiveness of therapy is the development of HIV drug resistance (DR).8 The development of resistance is associated with mutational processes occurring in the viral genome under the influence of various evolutionary factors.8
Virological failure and subsequent development of acquired DR is therefore frequently reported in certain countries.9-11 Since 2006, the WHO has recommended population-level investigations to assess and prevent pretreatment drug resistance (PDR) in low and low-middle income countries (LMICs); the majority of studies and surveys conducted to date indicate increasing levels of PDR, especially in sub-Saharan Africa.9, 12, 13 PDR in LMICs more often includes low levels of protease inhibitor (PI) mutation (<2%–3%), moderate levels of NRTI mutation (∼5%), and moderate to elevated levels of NNRTI mutation (10%).9 Moreover, several studies have reported increasing trends of PDR in LMICs, following ART rollout since the 1990s.14, 15 A recently conducted systematic review and meta-analysis, representing up to 56 044 adults in 63 countries, found that the prevalence of pretreatment NNRTI resistance is substantially rising annually, with PDR reaching: 23% (95% CI: 16%–29%) in southern Africa; 17% (95% CI: 5%–30%) in eastern Africa; 17% (95% CI: 6%–29%) in western and central Africa; 11% (95% CI: 5%–18%) in Latin America and the Caribbean; and 11% (95% CI: 2%–20%) in Asia.16
The Republic of Guinea is a located in West Africa. HIV prevalence in Guinea was approximately 1.6% in 2014. In 2019, it was practically unchanged, but treatment coverage also remains one of the lowest in the world, with less than a quarter (23%) of PLHIV on antiretroviral therapy (ART).3, 17 Among other things, Africa (especially Western and Central) has the highest diversity of HIV subtypes in the world. In West and Central Africa, the most common circulating recombinant form (CRF) is 02_AG. The A and G subtypes co-circulate in some countries, such as Nigeria.18-20 A 2016 study confirmed the prevalence of CRF 02_AG in Guinea.21
The Republic of Guinea is one of the countries affected by the Ebola epidemic. As such, there were significant difficulties in providing treatment to patients with HIV infection in the 2014–2015 period. This, in turn, may have affected the prevalence of DR. Therefore, more careful monitoring of HIV resistance is required in this region. However, the most recent data on DR in Guinea reported in the literature dates back to 2009, when the prevalence of primary DR was 8.9%.21 In 2016, it was not possible to analyze a sufficient number of samples to identify the prevalence of DR in the Republic of Guinea.22 In addition, in the above study, patients with an existing diagnosis were examined, but due to the low awareness of HIV infection in the Guinean population, patients who do not know about their infection can make a significant contribution to the genetic diversity of the virus.
2 METHODS
2.1 Study setting
During the study, 2168 blood plasma samples were obtained in 2015–2018 from apparently healthy people from the Republic of Guinea: blood donors and employees of UC RUSAL and their families. The clinical site serves the local population and provides routine blood testing and banking services. With the study's simple observational design, detailed demographic information about participants was not collected. Residents interacted with the site in relation to its routine regime of providing services. Employees of UC RUSAL and their families participated as part of worker healthcare support and protection. As part of overall safety precautions, only participants with a negative Ebola virus test were included. The surveyed persons indicated no known history of HIV infection. The study was conducted at the Center of Epidemiology and Microbiology in Kindia. Materials were collected by UC RUSAL employees and Institute of Applied Biological Research of Guinea specialists.
2.2 Participant enrollment
Inclusion criteria were: (i) ≥18 years of age; and (ii) no history of HIV infection.
2.3 Laboratory methods
Patient samples were tested for the presence of HIV antigens and antibodies to the virus by ELISA. Quantitative analysis of HIV RNA was carried out with a commercial kit, AmpliSens® HIV-Monitor-FRT (Central Research Institute of Epidemiology), with a sensitivity threshold of 500 copies/ml. Samples with a detectable viral load (VL) were analyzed using reverse transcription polymerase chain reaction (RT-PCR) and Sanger sequencing. For reverse transcription and amplification of HIV RNA, the RT-PCR-kit-Pro/Rev and PCR-kit-Pro/Rev commercial kits (Central Research Institute of Epidemiology) were used. Sequencing reactions were performed using the AmpliSens® HIVResist-Seq kit (Central Research Institute of Epidemiology) according to the instructions, as described earlier.23 For HIV genotyping, we used a 1302 nucleotide sequence spanning the pol gene (nt. 2253–3554). Coordinates given for the data represent the GenBank entry for HIV HXB2 (K03455.1). Analysis of sequence reaction products was performed using an ABI Prism 3500 genetic analyzer (Applied Biosystems).
Sequence data were analyzed using the NCBI Blast program to compare nucleotide sequences with those in the GenBank international database. Alignment of nucleotide sequences was executed with the MEGA (7.0) program using the ClustalW algorithm.24 For the construction of phylogenetic trees and subsequent phylogenetic analysis, the Neighbor-joining algorithm was used, which allows optimization of trees by the criterion of “balanced minimum evolution.” When assessing the reliability of phylogenetic relationships, we used multiple generations of samples using the bootstrap method for 1000 independent constructions of each phylogenetic tree.
Isolates were analyzed for recombination features using the REGA HIV-1 Subtyping Tool (3.0).25 Samples were also analyzed for phylogenetic relationships using MEGA and reference sequences from GenBank. This made it possible to more accurately assess the distribution of HIV-1 subtypes in the studied population. Analysis of sequences for the presence of DR mutations was performed using the Stanford University HIV DR Database.26
3 RESULTS
3.1 Study population
In total, materials were studied from 2168 patients. Their ages were within the range 18–58 years, with a median age of 38 years (β = 0.95, [37.58; 39.72]). Most of the examined patients were males (68.17% [β = 0.95; (66.21%; 70.13%]). Viral RNA was detected in 58 patients, aged 18–54 years. The median age was 36 years (β = 0.95, [32.53; 39.47]). The predominant age group was those from 31 to 45 years old (62.07%). The most represented in the sample were men. They made up 63.79% of the study group (β = 0.95, [52%; 76%]), while women represented 36.21% (β = 0.95, [24%; 49%]).
3.2 Molecular and serological HIV markers
Serological markers of HIV (anti-HIV antibodies, HIV antigen) were detected in 239 people, which is 11.02% (β = 0.95; [9.74%; 12.42%]) of the total sample. Note that 69.45% (β = 0.95; [63.19%; 75.23%]) of patients with identified HIV markers were young people aged 20–39 years. HIV RNA was detected in 58 people, which represents 24.27% (β = 0.95 [18.97%; 30.21%]) of patients in the seropositive group (2.68% of the total group).
3.3 HIV diversity and DR
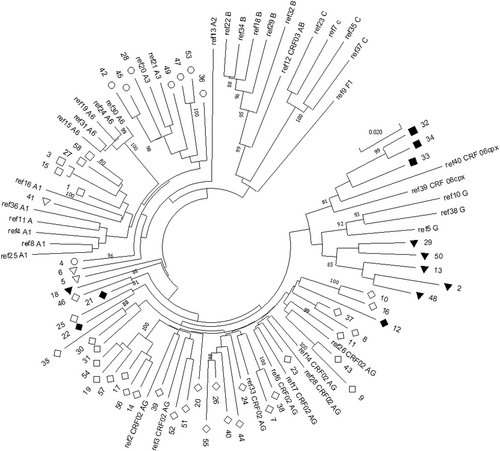
The HIV pol gene of all patients with a detectable VL was sequenced and submitted to GenBank (MT874291-MT874321, MT874310-MT874321, MT919401-MT919427). Analysis made it possible to identify the following HIV-1 subtype ratios. The circulating recombinant form (CRF), 02_AG (56.90% [β = 0.95; (43.23%; 69.84%)]), prevailed in the study group compared to: HIV A3 (13.79% [β = 0.95; (6.15%; 25.38%)]); A1 (12.07%; [β = 0.95; (4.99%; 23.30%)]); G (8.62% [β = 0.95; (2.86%; 18.98%)]); CRF_06cpx (5.17%; [β = 0.95; (1.08%; 14.38%)]); URF between A1 and G (1.72%; (β = 0.95; (0.04%; 9.24%)]); and A6 (1.72%; (β = 0.95; (0.04%; 9.24%)]) (Table 1, Figures 1 and 2).
| All patients | Patients with a detectable viral load | |
|---|---|---|
| Individuals | 2168 | 58 |
| Median age | 38 years (β = 0.95, [37.58; 39.72]) | 36 years (β = 0.95, [32.53; 39.47]) |
| Proportion men | 68.17% (β = 0.95; [66.21%; 70.13%]) | 63.79% (β = 0.95, [50.07%; 76.02%]) |
| Proportion women | 39.43% (β = 0.95; [37.36%; 41.52%]) | 36.21% (β = 0.95; [23.98%; 49.93%]) |

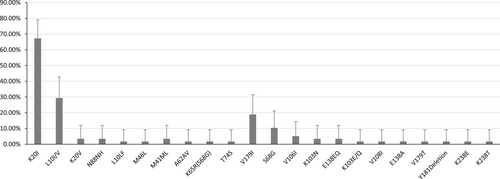
According to the analysis, 25% of patients had at least one significant mutation leading directly to HIV DR for their virus subtype. The mutations encountered cause resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors (Table 2, Figure 3). In one case, several mutations were encountered simultaneously, causing resistance to both classes of drug (M41ML + K65R + A62AV + S68G + K103N + V108I). Among them is the K65R mutation, which causes resistance to most NRTIs, together with the S68G mutation, which partially restores the replication defect associated with K65R.27 No major mutations associated with resistance to protease inhibitors were found.
| Sample name on phylogenetic tree (Figure 1) | Complete sample name | Subtype in REGA (version 3.0) | Subtype by phylogenetic analysis |
|---|---|---|---|
| 1 | 4352 | A1 | A1 |
| 2 | 6397 | G | G |
| 3 | 1128 | A1 | A1 |
| 4 | 3879 | A1 | A3 |
| 5 | 6436 | A1 | A1 |
| 6 | 4238 | A1 | A1 |
| 7 | 3169 | A1 | CRF 02AG |
| 8 | 1786 | CRF 02AG | CRF 02AG |
| 9 | 4886 | CRF 02AG | CRF 02AG |
| 10 | 2434 | CRF 02AG | CRF 02AG |
| 11 | 1912 | CRF 02AG | CRF 02AG |
| 12 | 5721 | Recombinant 02AG/A1 | CRF 02AG |
| 13 | 1286 | Recombinant G/A1 | G |
| 14 | 2492 | CRF 02AG | CRF 02AG |
| 15 | 1037 | HIV A | A6 |
| 16 | 1048 | CRF 02AG | CRF 02AG |
| 17 | 2356 | CRF 02AG | CRF 02AG |
| 18 | 1170 | Recombinant G/A1 | Recombinant G/A1 |
| 19 | 1156 | CRF 02AG | CRF 02AG |
| 20 | 1260 | CRF 02AG | CRF 02AG |
| 21 | 2709 | Recombinant 02AG/A1 | CRF 02AG |
| 23 | 2886 | CRF 02AG | CRF 02AG |
| 24 | 2595 | CRF 02AG | CRF 02AG |
| 25 | 2141 | CRF 02AG | CRF 02AG |
| 26 | 2046 | CRF 02AG | CRF 02AG |
| 27 | 1969 | A | A1 |
| 28 | 1976 | A | A3 |
| 29 | 2073 | G-like | G |
| 30 | 2813 | CRF 02AG | CRF 02AG |
| 31 | 2205 | CRF 02AG | CRF 02AG |
| 32 | 2501 | CRF 06CPX | CRF 06CPX |
| 33 | 2731 | CRF 06CPX | CRF 06CPX |
| 34 | 2279 | CRF 06CPX | CRF 06CPX |
| 35 | 2488 | CRF 02AG | CRF 02AG |
| 36 | 2130 | A1 | A3 |
| 37 | 2504 | CRF 02AG | CRF 02AG |
| 38 | 2505 | CRF 02AG | CRF 02AG |
| 39 | 2506 | CRF 02AG | CRF 02AG |
| 40 | 2526 | CRF 02AG | CRF 02AG |
| 41 | 2530 | A | A1 |
| 42 | 2547 | A | A3 |
| 43 | 2562 | CRF 02AG | CRF 02AG |
| 44 | 2564 | Recombinant 02AG/A1 | CRF 02AG |
| 45 | 1261 | A1 | A3 |
| 47 | 1215 | A1 | A3 |
| 48 | 1333 | Recombinant 14BG/G | G |
| 49 | 3816 | A | A3 |
| 50 | 2670 | G | G |
| 51 | 2412 | CRF 02AG | CRF 02AG |
| 52 | 2578 | CRF 02AG | CRF 02AG |
| 53 | 6778 | A | A3 |
| 54 | 1762 | CRF 02AG | CRF 02AG |
| 55 | 7032 | CRF 02AG | CRF 02AG |
| 56 | 1805 | CRF 02AG | CRF 02AG |
| 57 | 2309 | CRF 02AG | CRF 02AG |
| 58 | 954 | A | A1 |
| ref1_A3 | AB098332 | A3 | A3 |
| ref2_CRF02_AG | AB231896 | CRF 02AG | CRF 02AG |
| ref3__CRF02_AG | AB231898 | CRF 02AG | CRF 02AG |
| ref4_A1 | AB287376 | A1 | A1 |
| ref5_G | AF061641 | G | G |
| ref6_CRF02_AG | AF063224 | CRF 02AG | CRF 02AG |
| ref7_C | AF067155 | C | C |
| ref8_A1 | AF069670 | A1 | A1 |
| ref9_F1 | AF075703 | F1 | F1 |
| ref10_G | AF084936 | G | G |
| ref11_A | AF107771 | A | A |
| ref13_A2 | AF286237 | A2 | A2 |
| ref14_CRF02_AG | AF377954 | CRF 02AG | CRF 02AG |
| ref15_A6 | AF413987 | A6 | A1 |
| ref16_A1 | AF484509 | A1 | A1 |
| ref17_CRF02_AG | AY151001 | CRF 02AG | CRF 02AG |
| ref18_B | AY173951 | B | B |
| ref19_A6 | AY500393 | A6 | A6 |
| ref20_A3 | AY521629 | A3 | A3 |
| ref21_A3 | AY521631 | A3 | A3 |
| ref22_B | AY713409 | B | B |
| ref23_C | AY772699 | C | C |
| ref24_A6 | EF589043 | A6 | A6 |
| ref25_A1 | EU110087 | A1 | A1 |
| ref26_CRF02_AG | EU786671 | CRF 02AG | CRF 02AG |
| ref27_A1 | EU861977 | A1 | A1 |
| ref28__CRF02_AG | GU201514 | CRF 02AG | CRF 02AG |
| ref29_B | HM586190 | B | B |
| ref30_A6 | HQ161930 | A6 | A6 |
| ref31_A6 | HQ449397 | A6 | A6 |
| ref32_B | KJ771697 | B | B |
| ref33_CRF02_AG | KT124792 | CRF 02AG | CRF 02AG |
| ref34_B | M17449 | B | B |
| ref35_C | U46016 | C | C |
| ref36_A1 | U51190 | A1 | A1 |
| ref37_C | U52953 | C | C |
| ref38_G | U88826 | G | G |
| ref39_CRF_06cpx | MH605500.1 | CRF 06cpx | CRF 06cpx |
| ref40_CRF_06cpx | HQ529257.1 | CRF 06cpx | CRF 06cpx |
4 DISCUSSION
Table 3 There is a significant discrepancy between the number of HIV-positive people identified using ELISA and those who were found to have viral RNA. Moreover, the percentage of people in whom the virus was detected by the PCR method correlates with literature data on the prevalence of HIV infection in the Republic of Guinea. However, the significant percentage of apparently healthy people who are HIV seropositive suggests that the incidence of HIV may be higher than the literature suggests. To confirm this, additional screening studies with a confirmatory test are necessary.
| Mutation | Percentage in the group | Description of mutation |
|---|---|---|
| PI-resistance mutations | ||
| K20I | 67.24% | K20I is a PI-selected accessory mutation that reduces NFV susceptibility. |
| L10I/V | 29.31% | L10I/V are polymorphic, PI-selected accessory mutations that increase the replication of viruses with other PI-resistance mutations. |
| L10LF | 1.72% | L10F is a common nonpolymorphic, PI-selected accessory mutation associated with reduced susceptibility to DRV, FPV, IDV, LPV, and NFV. |
| M46L | 1.72% | M46I/L are relatively nonpolymorphic PI-selected mutations. In combination with other PI-resistance mutations, they are associated with reduced susceptibility to each of the PIs except DRV. |
| K20V | 3.45% | K20M/V are rare, relatively nonpolymorphic PI-selected mutations that have not been well studied. |
| N88NH | 3.45% | N88S is a nonpolymorphic mutation usually selected by NFV, ATV, or IDV which causes high-level resistance to NFV and ATV and low-level resistance to IDV and SQV. It increases susceptibility to FPV. N88D is a nonpolymorphic mutation selected by NFV usually in combination with D30N. N88H is a highly unusual mutation at this position. |
| NNRTI-resistance mutations | ||
| V179I | 18.96% | V179I is a polymorphic mutation that is frequently selected in patients receiving ETR and RPV. But It has little. if any. direct effect on NNRTI susceptibility. |
| S68G | 10.34% | S68G is a polymorphic mutation that is often selected in combination with K65R. It partially restores the replication defect associated with K65R. |
| V106I | 5.17% | V106I occurs in 1%–2% of viruses from untreated persons. It contributes to reduced NNRTI susceptibility in combination with other mutations. It has a weight of 1.5 in the Tibotec ETR genotypic susceptibility score despite not contributing much to reduced ETR susceptibility. It is commonly selected during DOR treatment in combination with mutations at position 227. |
| K103N | 3.45% | Causes a significant decrease in sensitivity to NVP and EFV |
| E138EQ | 3.45% | Reduces susceptibility to ETR and RPV by about two times |
| K103E/Q | 1.72% | Causes a significant decrease in sensitivity to NVP and EFV |
| V108I | 1.72% | V108I is a relatively nonpolymorphic accessory mutation selected in vitro and/or in vivo with each of the NNRTIs. It causes low-level reductions in susceptibility to NVP and DOR. Alone. it does not appear to reduce susceptibility to EFV, ETR or RPV. |
| E138A | 1.72% | Reduces susceptibility to ETR and RPV by about two times |
| V179T | 1.72% | V179T is a relatively rare nonpolymorphic mutation occasionally selected in patients receiving NNRTIs. It is associated with minimal. if any. reduction in ETR and RPV susceptibility. It has a weight of 1.0 in the Tibotec ETR genotypic susceptibility score. |
| Y181Deletion | 1.72% | Reduces susceptibility to NVP, ETR RPV and EFV by more than 50. 5. three and two times, respectively |
| K238E/T | 3.44% | K238T is a nonpolymorphic mutation selected in patients receiving NVP and EFV. It usually occurs in combination with K103N. It reduces susceptibility to NVP and EFV by about 5-fold. It may also reduce susceptibility to ETR and RPV. K238N is a nonpolymorphic accessory mutation that is also selected by NVP and EFV. It appears to have minimal. if any. effects on NNRTI susceptibility. |
| NRTI-resistance mutations | ||
| M41ML | 3.45% | M41L is a TAM that usually occurs with T215Y. In combination. M41L plus T215Y confer intermediate/high-level resistance to AZT and d4T and contribute to reduced ddI. ABC and TDF susceptibility. |
| A62AV | 1.72% | A62V is an accessory mutation that often occurs in combination with the multi-NRTI resistance mutations K65R or Q151M. A62V is widespread in subtype A viruses in former Soviet Union countries but A62 is otherwise nonpolymorphic. |
| K65R(S68G) | 1.72% | Causes medium/high resistance to TDF. ddI. ABC and d4T and low/medium resistance to 3TC and FTC. Increases sensitivity to AZT |
| T74S | 1.72% | L74V/I cause high-level resistance to ddI and intermediate resistance to ABC. L74S is a highly unusual mutation at this position. |
- Abbreviation: ABC, abacavir; ATV, atazanavir; AZT, azidothimidine; DDI, didanosine; DOR, doravirine; d4T, stavudine; DR, drug resistance; DRV, darunavir; EFV, efavirenz; ETR, etravirine; FPV, fosamprenavir; FTC, emtricitabine; IDV, indinavir; LPV, lopinavir; NFV, nelfinavir; NVP, nevirapine; RPV, rilpivirine; SQV, saquinavir; TDF, tenofovir; 3TC, lamivudine.
The genetic diversity identified is consistent with the literature on HIV subtypes circulating in West Africa and the high genetic diversity of HIV in Africa in general. In the overwhelming majority of cases, the circulating recombinant form, 02_AG, is found. In this situation, special attention should be paid to isolates, the genotyping of which showed that more refined analysis (i.e., of recombination) is necessary. To gain a more complete picture, we additionally performed phylogenetic analysis together with isolates obtained in the 2016 study (Figure 4). On both phylogenetic trees, sample 18 was isolated, subtyped in REGA 3.0 as a recombinant of genotypes A1 and G, but not belonging to CRF02_AG (Figure 3). It is interesting to note that it forms a cluster with the isolate submitted under the number LT976766, which was assigned to CRF02_AG in 2016; this can be explained by the lack of information on the protease gene sequence in this sample. It is important to note that samples 44 and 46, identified in REGA 3.0 as unknown recombinant forms, are most closely clustered with CRF02_AG in both phylogenetic trees. It should be noted that samples 44, 46, and 13, according to the results of REGA HIV-1 analysis, are also recombinants between the known CRF_02AG and A1 subtypes (Figure 3).

Based on the analysis, we assume a significant contribution of various recombinant HIV forms to the genetic diversity of the virus in the studied region. In this regard, it is extremely important to study whole HIV genomes in the Republic of Guinea to identify all recombinant forms present and the most common recombination points. Since accurate genotyping is critical to screening a sample set for resistant HIV variants, more research is needed to assess the contribution of recombinant forms to the genetic diversity of the virus in the region. Insufficient attention to the high diversity of HIV recombinants, and the lack of complete data on common points of recombination, can lead to an erroneous determination of the presence or absence of viral.
The incidence of mutations associated with HIV resistance to ARVs was relatively high. Given the increasing number of patients starting ART, the high incidence of primary DR can lead to frequent cases of treatment failure and consequent changes in treatment. To prescribe effective treatment regimens, it is necessary to introduce studies on the presence of primary resistance in newly diagnosed patients.
A similar, relatively high incidence of DR mutations, in untreated infected individuals, has been shown in Sierra Leone. In 2014–2016, an Ebola outbreak affected Sierra Leone and its neighbors, including the Republic of Guinea. During this time period, medical services were disrupted in the region, including HIV care. However, a lower number of resistant HIV-1 forms were seen in Sierra Leone according to analysis.28
The 24 mutations identified in naive patients here would most likely have approximately the same frequency of occurrence in a larger sample set. This is to be expected as there is no pressure on the virus from antiretroviral drugs in the study group, and mutations spontaneously arise and disappear in generations of the virus.
It is interesting to note that, in more than half of the cases (72%), there were substitutions at the twentieth position (pol gene). It can be seen that, of these mutations, K20I stands out the most. It is a consensus variant for subtypes CRF_02AG and G, but reduces sensitivity to nelfinavir in subtypes B and C. However, there is evidence that this mutation may enhance viral replication in non-B subtypes.29 This mutation had a significantly higher incidence than all the others (67.24% [β = 0.95, (53.66%; 78.99%]), despite the absence of selection factors in the form of ART; this may be a consequence of an advantage received by carriers (virus) of this mutation (Figure 4).
A nonpolymorphic variant of K20V has also been identified, which is rare, and its effect on HIV susceptibility to ARVs is poorly understood. Also, two mutations at the tenth position were identified. One of which, detected in one case (L10LF), is a minor mutation of resistance to PI. The other, L10I, was found in 29.03% of cases and increases the replication of viruses with other resistance mutations to SP.30 Two rare mutations were identified: M46M_M and N88NH. Mutations at these positions have been associated with resistance to protease inhibitors. These variants, however, have not been described among them.
Among the mutations associated with HIV resistance to nonnucleoside reverse transcriptase inhibitors, a relatively rare, nonpolymorphic V179T mutation has been shown, which is sometimes selected for in patients receiving NNRTIs. This is due to a minimal decrease in the sensitivity to etravirine and rilpivirine.29, 31 Several substitutions were also encountered at position 238: K238T, which reduces susceptibility to nevirapine and efavirenz by about 5-fold; K238R, which is a common polymorphism that does not reduce susceptibility to NNRTIs32; and K238E, a rare mutation at this position, the effect of which is not well described in the literature.
The identified mutations associated with DR, both arising spontaneously and as a result of transmission by patients with treatment-resistant infections, can actively spread among new patients due to: the low awareness of the population about HIV infection; and inaccessibility of medical care and contraception. A consequence of low awareness of blood-borne infections is also an increased risk of trauma-based transmission of the virus, the possibility of which has been shown in various studies.33
In conclusion, we note that even a joint analysis, with data obtained in the course of this study and in 2016, was insufficient to fully clarify the genetic diversity of HIV in the Republic of Guinea. More detailed studies, on larger sample sets, are needed to elucidate the genetic profile and etiological structure of the virus in a territory with such a complex epidemiological situation. One aspect is clear however. The detection of HIV-1 mutations associated with DR, in individuals who have never received ART, is a cause for concern. It suggests that: new infections are occurring with strains that already have resistance; and the expansion of resistance is not always directly associated with selective drug pressure. Among the likely reasons for the high prevalence of primary HIV DR in the Republic of Guinea, drug availability is probably the key. The consequence of this is the lack of adherence of patients to treatment, the formation and transmission of resistant variants of the virus in the population.
AUTHOR CONTRIBUTIONS
Alexander N. Shchemelev: Conducting research; literature selection; statistical data processing; illustrative material preparing, article text writing. Sanaba Boumbaly: Planning and organization of research, sample delivery. Yulia V. Ostankova: Conducting research; literature selection; statistical data processing; illustrative material preparing; article text writing. Elena B. Zueva: Conducting research. Alexander V. Semenov: General guidance; article text writing. Areg A. Totolian: General guidance; article text writing.
ACKNOWLEDGMENT
Rospotrebnadzor (Federal Service for Supervision of Consumer Protection); and the program of Russian-Guinean cooperation on the implementation of the order of the Government of the Russian Federation, dated December 22, 2017 No. 2409-r.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Data available on request from the authors.




