Bioinformatics analysis of the s2m mutations within the SARS-CoV-2 Omicron lineages
Now more than 2 years into the COVID-19 pandemic, the detailed mechanisms of the SARS-CoV-2 viral life cycle have yet to be elucidated. The 5ʹ- and 3ʹ-untranslated regions (UTRs) of the RNA genome are of particular interest as potential targets for antiviral therapies, given that they contain structural elements predicted to aid in the viral life cycle.1 Located within the 3ʹ-UTR, the stem-loop II motif (s2m) is a conserved structural element that has been hypothesized to aid in viral transcription or RNA silencing pathways beneficial to the virus.2 In SARS-CoV, the virus responsible for the 2002–2003 SARS outbreak, s2m-deficient coronaviruses have subtle differences in their potency; however, the role of the s2m regarding viral fitness is yet to be uncovered.3 In addition to these proposed functions, other coronaviruses' UTRs have been shown to hijack host regulatory machinery, such as microRNAs (miR).4 We have recently shown that the SARS-CoV-2 s2m forms homodimeric kissing complexes converted to a stable duplex structure by the viral nucleocapsid (N) protein, and that it harbors two binding sites for the host miR-1307-3p, which has been suggested to regulate various cytokines and their receptors, such as IL18 and IL6R.5 These interactions highlight the potential for the s2m to participate in essential events of the viral life cycle, including recombination events and viral hijacking of host biomolecules.
The emergence of the SARS-CoV-2 Omicron variant (B.1.1.529 + BA.*) has shifted interest to viral fitness based on genetic and phenotypic differences. Omicron, when compared to the Delta (B.1.617.2 + AY.*), Beta (B.1.351), and Alpha (B.1.1.7) variants, contains several mutations across the spike S protein and accessory proteins that aid in transmissibility, immune evasion, tropism shift, and viral entry into the cell.6 The BA.1* sublineages, known as the original Omicron variant, surged in January 2022 and were outcompeted in March of 2022 by BA.2* (represented by mostly sublineages BA.2, BA.2.9, and BA.2.12.1). By June 2022, the BA.4, BA.5, and BA.5.1 sublineages have shown dominance over the BA.2 sublineages, and currently represent most new Omicron cases worldwide, suggesting further increased fitness. Thus, in this study we performed a bioinformatics analysis monitoring s2m sequence changes among Omicron sublineages, as they may have the potential influence viral fitness.
SARS-CoV-2 genomes for the Omicron and Alpha sublineages were collected from the GISAID EpiCoV database in the FASTA file format for cases submitted through June 2022, which were selected based on PANGO designated lineages.7 All lineages denoted by a “*” indicate complete inclusion of sublineages under the designation. The Omicron sublineages BA.2, BA.2.9, BA.2.12.1, BA.4, BA.5, and BA.5.1 were chosen due to their high prevalence within all Omicron cases, and for comparison, we have also analyzed the Alpha variant. The hCoV-19 Wuhan SARS-CoV-2 virus (GISAID: EPI_ISL_402123) was used as a reference genome.8 Sequence analysis of the s2m element was performed using a custom R script utilizing the following packages: BiocManager, Biostrings, DECIPHER, hiReadsProcessor, adegenet, stringr, and ape. We converted the SARS-CoV-2 reference sequence to a FASTA file and added it to each batch, which contained less than 10,000 sequences. The first 29,000 nucleotides of each sequence were removed to reduce computational time, aligning the remaining nucleotides at the 3ʹ end. According to GISAID, sequences containing less than 29,000 nucleotides are considered “incomplete.” These sequences, which are a source of error in this analysis, were identified by our script, removed, and accounted for in the “removed sequences” count. Additionally, any sequences which contain single nucleotide insertions or large deletions within the remaining nucleotides were also removed due to their disruption of the alignment and included in the “removed sequences” count. The remaining sequences were then realigned. For the Alpha variant, from November 2020 to December 2021 a total of 1,091,684 sequences were analyzed and 125,080 were removed (11.46%). For Omicron, from January to June 2022, 2,764,678 sequences were analyzed, and 25,625 sequences were removed (0.93%). The s2m was isolated from the trimmed sequences, where mutations and corresponding metadata (accession ID, geographic location, and collection date) were output in a CSV file. Subsequently, s2m sequences were identified that were an exact match to the wild-type s2m, (TTCACCGAGGCCACGCGGAGTACGATCGAGTGTACAGTGAA), or to the 26-nucleotide deletion within the s2m (TTCACC--------------------------TACAGTGAA), named “Δ(7-32) s2m.” Additionally, sequences in which the 3ʹ-end of the deletion is translocated (TTCACCTA--------------------------CAGTGAA), or in which the Δ(7-32) appears as “N” were identified as the Δ(7-32) s2m. Further sequences where the s2m was truncated at the 7th position (TTCACC-----------------------------------), identified as the Δ(7-41) s2m, or which are truncated before the s2m, identified as “truncated before s2m” (TBs2m), were also accounted for and reported.
The wild-type s2m and Δ(7-32) s2m sequences were purchased from Dharmacon Inc. and resuspended in 10 mM cacodylic acid, pH 6.5. One-dimensional and 1H-1H NOESY NMR spectroscopy were used to assign the G and U imino proton resonances of the Δ(7-32) s2m.
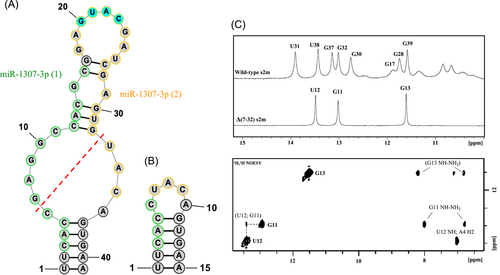
We found that BA.1* sublineages contained the wild-type s2m in high prevalence, whereas the Δ(7-32) s2m is predominant within the BA.2* sublineages. This deletion mutation eliminated the upper stem of the s2m, specifically, nucleotides 29,734 through 29,759 of the SARS-CoV-2 reference genome. The RNAstructure software predicts that the lower stem of the wild-type s2m remains intact in the Δ(7-32) s2m (Figure 1A,B) and our 1H NMR spectroscopy results verified this prediction. Only three resonances are present in the imino proton resonance region of the Δ(7-32) s2m (Figure 1C, top panel), with two of them having the same chemical shifts as the resonances previously assigned to U38 and G39 in the wild-type s2m lower stem.5, 9 To assign the Δ(7-32) s2m imino proton resonances, we performed a 1H-1H NOESY (Figure 1C, lower panel). The resonance at 13.4 ppm was assigned to the U12 imino proton based on its cross-peak with the A4 H2 proton, and the resonances at 12.9 and 11.5 ppm were assigned to G imino protons based on the two strong NOE cross-peaks with their own NH2 protons. The U12 imino proton is expected to have a stronger cross-peak with the G11 imino proton than with the G13 imino proton since it is 3.5 Å from G11 and 5.0 Å from G13 imino proton, respectively.10 Thus, the observed cross-peak between the U12 imino and the imino proton resonance at 12.9 ppm allows the assignment of this resonance to the G11 imino proton. Like in the wild-type s2m,5, 9 the imino proton resonances of U1 and U2 are not observable due to base pair fraying.
We found that the Δ(7-32) s2m was present in a small percentage of BA.1* (11.1%) and BA.1.1* (4.43%) cases worldwide before the appearance of BA.2*. While the Δ(7-32) s2m has been previously identified in an early BA.1/BA.2 recombinant variant,11 our analysis of the dominant BA.2* sublineages reveals that both BA.2 and BA.2.9 have an increased prevalence of the Δ(7-32) s2m from January to June 2022. BA.2 and BA.2.9 start at 51.1% and 59.4%, respectively, in January, and by the end of June 2022 these percentages rose to 79.5% for BA.2% and 84.6% for BA.2.9 (Figure 2A, green and blue).
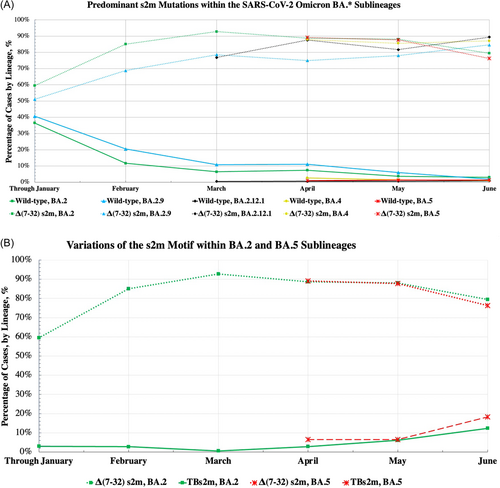
The BA.2.12.1 sublineage, which spread significantly in March 2022, had a Δ(7-32) s2m prevalence of 83.4% and then 89.5% by the end of June of 2022 (Figure 2A, black). We also analyzed the Omicron BA.4, BA.5, and BA.5.1 sublineages: BA.4 starts at 87.9% and BA.5 at 89.2% for Δ(7-32) s2m in April 2022 with these percentages changing to 87.0% and 76.3%, respectively, by June 2022 when cases rose significantly (Figure 2A, yellow and red). The number of cases for the BA.5.1 sublineage is only significant in June 2022 and the Δ(7-32) s2m is present at 89.3%. Although the Δ(7-32) s2m percentage for BA.5 decreases in June 2022, this is accompanied by an increase in the percentage of the TBs2m sequences, which are terminated before the s2m, and thus are a source of uncertainty in our analysis (Figure 2B, red). A similar trend is observed for BA.2, the only other sublineage which shows an apparent decrease of the Δ(7-32) s2m percentage in June 2022 as compared to the previous month (Figure 2B, green). The TBs2m percentages increase from March to June 2022 across all the Omicron sublineages, ranging between 0.56% and 18.3% with an average of 7.46%. Thus, BA.2*, BA.4, and BA.5* show a high percentage of Δ(7-32) s2m throughout the February–June 2022 period. Interestingly, our analysis of the Δ(7-32) s2m also revealed that in each Omicron sublineage, an increasing percentage of s2m sequences lack the rest of the sequence past position 7 (Δ[7-41] s2m) which ranges between 0.2% and 10.4%, with an average of 3.28% of cases.
To determine if the Δ(7-32) s2m and Δ(7-41) s2m are unique to the Omicron sublineages, we analyzed the Alpha variant, which is the closest predecessor of Omicron.12 The full-length wild-type s2m was retained in 96.0% of the Alpha variant from November 2020–December 2021, and no Δ(7-32) s2m or Δ(7-41) s2m were identified in this variant. The TBs2m percentage was 3.67% for the Alpha variant. We noticed that in Alpha, the percentage of removed sequences ranges from 1.34% to 27.46% during November 2020 to December 2021, with a spike in December, averaging to 11.46%. This is in contrast to the removed sequence count of Omicron, which ranged from 0.29% to 1.82% throughout January to June 2022 and averaged 0.93%. We attribute the increase in removed sequences for Alpha to a drop in sequence quality, reflective of incomplete sequences submitted.
The rise of the Δ(7-32) s2m mutant as the dominant s2m phenotype suggests that this mutation may influence the overall fitness of the SARS-CoV-2 Omicron variant. An alternative explanation is that the deletion mutant has increased in frequency by hitchhiking on a positively selected variant elsewhere in the genome. Such a large removal of the sequence could provide either a gain or loss of function to the s2m, altering its predicted functions in miR binding or dimer formation. The Δ(7-32) s2m mutation removes the terminal loop palindromic sequence and the miR-1307-3p binding sites (Figure 1), both of which were previously identified and hypothesized to aid in the viral life cycle.5 Thus, we predict that the Δ(7-32) s2m has reduced miR-1307-3p binding interactions and dimer formation when compared to the wild-type s2m. Experiments are currently in progress in our laboratory to elucidate the role of the Δ(7-32) s2m in the context of the SARS-CoV-2 3ʹ-UTR and whether this affects long-range RNA-RNA interactions. Further work is necessary to determine if this s2m mutant indeed confers a selective advantage to Omicron. We also identified the Δ(7-41) s2m which could be part of a larger truncation within the 3ʹ-UTR. As SARS-CoV-2 continues to evolve, we emphasize the s2m for future monitoring and highlight the motif as a target of interest regarding viral fitness for both existing and future SARS-CoV-2 variants and lineages.
AUTHOR CONTRIBUTIONS
Mihaela Rita Mihailescu, Jeffrey D. Evanseck and Patrick E. Lackey were involved in planning and supervised the work, Caleb J. Frye and Mihaela Rita Mihailescu conceived of the presented idea, Morgan Shine and Joseph A. Makowski wrote and modified the bioinformatics script used for analysis. Caleb J. Frye performed the bulk of the bioinformatics analysis with help from Adam H. Kensinger, Caylee L. Cunningham and Ella J. Milback and Mihaela Rita Mihailescu and Caylee L. Cunningham performed the experimental NMR spectroscopy experiments. Caleb J. Frye wrote the initial manuscript and all authors discussed the results and contributed to the final revised manuscript.
ACKNOWLEDGMENT
This research was supported by NSF CHE-2029124 RAPID (M. R. M., J. D. E.), NSF MRI Supercomputer CHE-1726824, and NSF CHE-1950585 REU (M. S., P. E. L., J. D. E.) grants.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA AVAILABILITY STATEMENT
All data is available upon request to the corresponding author.




