Dissecting the potential role of hepatitis E virus ORF1 nonstructural gene in cross-species infection by using intergenotypic chimeric viruses
Debin Tian and Danielle M. Yugo contributed equally to this work.
Abstract
Hepatitis E virus (HEV) infects humans and more than a dozen other animal species. We previously showed that open reading frame 2 (ORF2) and ORF3 are apparently not involved in HEV cross-species infection, which infers that the ORF1 may contribute to host tropism. In this study, we utilize the genomic backbone of HEV-1 which only infects humans to construct a panel of intergenotypic chimeras in which the entire ORF1 gene or its functional domains were swapped with the corresponding regions from HEV-3 that infects both humans and pigs. We demonstrated that the chimeric HEVs were replication competent in human liver cells. Subsequently, we intrahepatically inoculated the RNA transcripts of chimeras into pigs to determine if the swapped ORF1 regions confer the chimeras’ ability to infect pigs. We showed that there was no evidence of infectivity in pigs for any of the chimeras. We also investigated the role of human ribosome protein sequence S17, which expanded host range in cultured cells, in HEV cross-species infection. We demonstrated that S17 insertion in HEV ORF1 did not abolish HEV replication competency in vitro, but also did not expand HEV host tropism in vivo. The results highlight the complexity of the underlying mechanism of HEV cross-species infection.
Highlights
-
Four intergenotypic chimeric HEVs in the backbone of HEV-1 containing ORF1 or its functional domains from HEV-3 were replication-competent in vitro.
-
A recombinant HEV in the backbone of HEV-1 with insertion of human ribosome protein sequence S17 in the ORF1 was replication-competent in cultured liver cells.
-
The four chimeric HEVs and the recombinant HEV failed to experimentally infect pigs.
-
The ORF1 alone is probably not involved in HEV host tropism.
1 INTRODUCTION
Hepatitis E virus (HEV) is the leading cause of acute viral hepatitis E worldwide. In industrialized countries, clinical cases of hepatitis E are sporadic and mainly transmitted zoonotically via direct contact with infected animals or via consumption of undercooked animal meat products.1 In many low-income countries, HEV infection is endemic or epidemic and occasionally associated with large outbreaks.2-4 It is estimated that approximately 20 million HEV infections and 3.3 million cases of acute hepatitis E occur annually worldwide.5 In addition to acute hepatitis E, chronic HEV infection has also been well documented in immunosuppressed individuals, such as organ transplant recipients.6, 7 Furthermore, a high mortality rate has been reported in HEV-infected pregnant women.8 Recently, HEV-related neurological sequelae, such as Guillain-Barré syndrome and neuralgic amyotrophy, as well as renal disease, such as membranoproliferative glomerulonephritis and cryoglobulinemia have become significant clinical problems.9-12
HEV belongs to the family of Hepeviridae which consists of two distinct genera.13 The genus Orthohepevirus is subdivided into four species, Orthohepevirus A-D. The species Orthohepevirus A includes at least eight different HEV genotypes, HEV-1 to HEV-8, infecting humans and a plethora of other animal species.14, 15 HEV-1 and HEV-2 are restricted to humans under natural circumstances, but can infect nonhuman primates under experimental conditions.5, 16 HEV-3 and HEV-4 are zoonotic and can cross species barriers infecting humans and other animal species including pigs and rabbits.16, 17 HEV-5 and HEV-6 have been identified in wild boars, but HEV-5 has also been shown to infect nonhuman primates experimentally.18-20 HEV-7, initially identified from a dromedary camel and a human organ transplant recipient, can infect nonhuman primates under experimental conditions.21 HEV-8 was identified from a bactrian camel, and can also infect nonhuman primates, and therefore likely also humans.22, 23 More recently, it has been reported that rat HEV from species Orthohepevirus C also infects humans.24, 25
HEV is a single-stranded, positive-sense, RNA virus.26 The viral genome of approximately 7.2 kb in size is comprised of a 5′-cap structure followed by a short 5′-untranslated region (5′-UTR), three partially-overlapping major open reading frames (ORFs), and a short 3′-UTR followed by poly (A) tail.14, 26 The ORF1 encodes a viral nonstructural polyprotein. Multiple functional domains within ORF1 have been identified including a methyltransferase, papain-like cysteine protease, helicase, and RNA-dependent RNA polymerase.27-29 Additionally, the ORF1 also contains three domains with largely unknown function: a Y domain, a proline-rich hypervariable region (HVR) with a high level of intra- and inter-genotypic sequence diversity, and an adjacent X domain.30-32 The ORF2 encodes the immunogenic capsid protein that induces neutralizing antibodies and is an important vaccine target.14 The ORF3, which overlaps ORF2, encodes a small multifunctional protein that is involved in virus replication.33, 34 A novel small ORF4, which is translated under endoplasmic reticulum stress, has been identified only in HEV-1.35
The underlying viral or host factors that determine HEV host range remain unknown. Dissection of the HEV viral attachment protein which interacts with the host cellular receptor will greatly help understanding the mechanism of HEV cross-species infection. In our previous studies, we investigated the potential role of the ORF2 gene in HEV cross-species infection, since the ORF2 codes for the capsid protein which presumably is the viral attachment protein that binds to host cell receptor.36 We constructed intergenotypic chimeric HEVs by swapping viral ORF2 and ORF3 genes between HEV-1 which infects only humans and HEV-3 which infects both humans and pigs, also between HEV-1 and HEV-4 which infects both humans and pigs, and subsequently tested the infectivity of the chimeras in pigs.37, 38 Our results showed that the chimeric viruses with swapped ORF2/ORF3 genes failed to expand its host range, suggesting that the ORF2/ORF3 genes are likely not involved in HEV host range determination, thus inferring that ORF1 may play a role in HEV cross-species infection.37, 38 This inference is further supported by the demonstration that insertion of the human ribosomal protein sequence S17 in the ORF1 HVR of HEV enhanced virus replication and expanded the recombinant virus's ability to infect cell lines of different animal origins in vitro.39, 40
Therefore, in this present study, we hypothesized that the ORF1 gene plays a role in HEV host tropism. To test the hypothesis, we utilized an infectious complementary DNA (cDNA) clone of HEV-1 (human Sar-55 strain, only infecting humans) as the genomic backbone to construct a panel of intergenotypic chimeric HEVs by swapping with different ORF1 regions of zoonotic HEV-3 (infecting both pigs and humans). Additionally, we also constructed a recombinant virus in which we cloned a 171 base pair (bp) of the human ribosomal protein sequence S17 in the HVR of HEV-1 genomic backbone. The chimeric constructs were tested in vitro for replication competency, and subsequently in pigs for infectivity.
2 MATERIALS AND METHODS
2.1 Cells and hepatitis E virus infectious complementary DNA clones
A subclone of the human hepatoma Huh7 cell line, Huh7-S10-3, a gift from Dr. Suzanne U. Emerson (National Institute of Health, Bethesda, MD) was used in this study. The cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) at 37°C with 5% CO2.41 The HEV infectious cDNA clone pSK-HEV-2, which was derived from the genotype 1 human HEV Sar-55 strain (HEV-1), was also kindly provided by Dr. Suzanne U. Emerson.42 A genotype 3 swine HEV infectious cDNA clone pSHEV3 (HEV-3), which was derived from swine HEV Meng strain, was produced and used from our previous study.43
2.2 Construction of chimeric HEV clones
To construct intergenotypic chimeric HEV clones, the HEV-1 infectious cDNA clone (pSK-HEV-2) was first modified by introducing three different restriction enzyme sites. By using overlap polymerase chain reaction (PCR), Mfe I (5′-CAATTG-3′), Fse I (5′-GGCCGGCC-3′), and Avr II (5′-CATTGG-3′), were inserted into the flanking regions of HVR and X domains (Figure 1). The modified HEV-1 clone was designated as SKHEV2-MFA, and used as the backbone for the subsequent constructions of chimeric HEV clones (Figure 1).
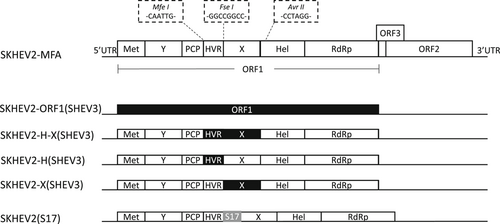
To produce the chimeric HEV clone SKHEV2-ORF1(SHEV3), the entire HEV genomic sequence, including the ORF1 that is derived from HEV-3 Meng strain clone pSHEV3, and the ORF2 and ORF3 that are derived from clone SKHEV2-MFA, was custom synthesized (GenScript, Piscataway, NJ). The genomic fragments coding for the HVR-X region, HVR region, X domain were amplified by PCR from HEV-3 clone pSHEV3 and swapped into the genomic backbone clone of HEV-1 SKHEV2-MFA via appropriate restriction enzymes sites. The final chimeric HEV clones were designated as SKHEV2-H-X(SHEV3), SKHEV2-H(SHEV3), and SKHEV2-X(SHEV3), respectively (Figure 1). Additionally, a human ribosomal protein S17 coding sequence (171 bp) derived from the HEV-3 Kernow C-1 P6 strain39 was cloned into the HVR of the HEV-1 backbone SKHEV2-MFA to produce the recombinant clone SKHEV2(S17) (Figure 1). All of the chimeric constructs were confirmed by Sanger DNA sequencing.
2.3 In vitro RNA transcription
The plasmid DNAs of each chimeric HEV construct and its parental infectious clone were linearized with Bgl II for HEV-1 SKHEV2-MFA backbone and chimeric clones, or with Xba I for HEV-3 pSHEV-3 infectious clone. Full-length capped genomic RNA transcripts for each of the parental and chimeric HEV clones were transcribed using mMessage mMachine T7 kit (Ambion), as described previously (37). The capped full-length genomic RNA transcripts were used for in vitro transfection of Huh7-S10-3 cells to determine the replication competency of the chimeric viruses, or immediately diluted in three volumes of cold RNase/DNase/proteinase-free phosphate-buffered saline (PBS) and frozen on dry ice in aliquots for use the next day for intrahepatic inoculation of pigs to determine virus infectivity.
2.4 In vitro transfection
Full-length capped genomic RNA transcripts were transfected into Huh7-S10-3 cells in six-well plate using DMRIE-C reagent (Invitrogen) according to the manufacturer's instruction. The transfected cells were cultured in DMEM maintenance medium with 4% FBS at 34.5°C. About 3 days after transfection, the cell monolayer was detached by treatment with 2.5% Trypsin-EDTA. DMEM maintenance medium was subsequently added to the suspended cells, which were then transferred into a 48-well culture plate and incubated at 34.5°C for another 3 days before testing for evidence of virus replication by a HEV-specific immunofluorescence assay (IFA).
2.5 Immunofluorescence assay
The IFA was conducted as previously described.43 Briefly, the transfected cells in the cell culture plate were fixed with cold acetone, washed with PBS and incubated with chimpanzee 1313 anti-HEV convalescent-phase serum.44 Cells were rinsed with PBS and stained with fluorescein isothiocyanate-conjugated goat anti-human immunoglobulin G (IgG) (Molecular Probes). The stained cells were washed by PBS and viewed under a fluorescence microscopy.
2.6 Intrahepatic inoculation of capped HEV genomic RNA transcripts into the liver of pigs
To test for the infectivity of the chimeric HEVs, we intrahepatically inoculated the capped full-length genomic RNA directly into the livers of pigs (Table 1) to bypass the in vitro cell culture system which is insufficient for HEV propagation. Briefly, 4-week-old HEV-seronegative specific-pathogen-free pigs were divided into eight groups (groups 1-8) with three pigs per group, and housed separately for each group in a BSL-2 swine research facility. The animal study was approved by the Institutional Animal Care and Use Committee and the Institutional Biosafety Committee at Iowa State University (approval numbers IACUC-19-327 and IBC-19-215). Under anesthesia, each pig was inoculated by a board certified veterinary internist, intrahepatically, using an ultra-sound guided technique into four different sites of each liver (~200 μL capped RNA transcripts per site). The three pigs in group #1 were intrahepatically inoculated with 0.8 mL of PBS buffer as a negative control. Pigs in groups 2 to 8 were intrahepatically inoculated with 0.8 mL capped HEV genomic RNA transcripts from each of the chimeric or parental clones, respectively. The capped RNA transcripts of a HEV-3 swine HEV infectious clone SHEV3 (group 3 pigs) was used as a positive control. The inoculated animals in each group were monitored for 10 weeks postinoculation (wpi) for evidence of HEV infection. Fecal and serum samples were collected before inoculation and weekly thereafter from each pig. At necropsy, serum, bile, and samples of liver tissue were also collected from each pig and stored at −80°C.
| Group | No. of pig | Inocula | Genomic backbone (genotype) | Donor sequence in the swapped gene (genotype) |
|---|---|---|---|---|
| 1 | 3 | PBS | NA | NA |
| 2 | 3 | SKHEV2-MFA | Sar-55 (HEV-1) | NA |
| 3 | 3 | SHEV3 | Meng (HEV-3) | NA |
| 4 | 3 | SKHEV2(S17) | Sar-55 (HEV-1) | Ribosomal protein sequence S17 |
| 5 | 3 | SKHEV2-ORF1(SHEV3) | Sar-55 (HEV-1) | ORF1 (HEV-3) |
| 6 | 3 | SKHEV2-H-X(SHEV3) | Sar-55 (HEV-1) | HVR and X domain (HEV-3) |
| 7 | 3 | SKHEV2-H(SHEV3) | Sar-55 (HEV-1) | HVR domain (HEV-3) |
| 8 | 3 | SKHEV2-X(SHEV3) | Sar-55 (HEV-1) | X domain (HEV-3) |
- Abbreviations: HEV, hepatitis E virus; HVR, hypervariable region; NA, not applicable; ORF1, open reading frame 1; PBS, phosphate-buffered saline.
2.7 Nested reverse transcription-polymerase chain reaction for detection of HEV RNA
Viral RNAs from weekly serum samples were isolated by using the Viral RNA isolation kit (Zymo Research) according to the instructions of manufacturer. Viral RNAs from weekly fecal samples, as well as from liver and bile samples collected at necropsy were isolated using TRIzol LS (Zymo Research) according to the manufacturer's instruction. The extracted RNAs were used for cDNA synthesis using SuperScript IV First-Strand Synthesis System (Invitrogen) following the manufacturer's instruction. The synthesized cDNAs were then used to amplify HEV gene-specific fragments by a nested RT-PCR using DreamTaq Hot Start PCR Master Mix (Thermo Fisher Scientific). The primers used for cDNA synthesis and reverse transcription-PCR (RT-PCR) amplification (Table S1) are designed as HEV strain-specific. The sensitivity of the nested RT-PCR assay was <10 copies per reaction. The amplified PCR products in the second round PCR were visualized by gel electrophoresis on 1% agarose in Tris-acetate-EDTA buffer (Thermo Fisher Scientific). The HEV RNA transcripts from parental clones of SHEV3 (HEV-3) and SKHEV2-MFA (HEV-1) were used as positive control and nuclease-free water as negative control in the cDNA synthesis.
2.8 Enzyme-linked immunosorbent assay to detect immunoglobulin G anti-HEV antibodies in inoculated pigs
An enzyme-linked immunosorbent assay (ELISA) was utilized to test for the presence of IgG anti-HEV antibodies in serum samples as described previously.45, 46 Briefly, an immunodominant region of HEV capsid ORF2 protein (amino acids 452-617) was used as the ELISA antigen (GenWay Biotech Inc, San Diego, CA) to coat a 96-well plate. The antigen-coated plates were incubated with each serum sample diluted in 1:100 blocking buffer. Pre-immune and hyperimmune swine serum samples were included as the negative and positive controls, respectively. Horse-radish peroxidase-conjugated goat anti-swine IgG (Sigma-Aldrich) was used as the secondary antibody at 1:2000 dilution in blocking buffer. The plate was developed using TMB Microwell Peroxidase Substrate system (SeraCare, Milford, MA) and read at 450 nm on a Promega GloMax Discover machine. The ELISA cut-off value was calculated as the average “P” value per duplicate sample divided by the average “N” value of the duplicate negative control samples with values above 2.0 considered positive.
3 RESULTS
3.1 The HEV-1-based intergenotypic chimeric clones and the recombinant clone SKHEV2(S17) with S17 insertion are replication competent in human liver cells in vitro
To investigate if the HEV ORF1 regions are involved in host tropism, four intergenotypic chimeric clones between HEV-1 and HEV-3 were constructed: SKHEV2-ORF1(SHEV3), SKHEV2-H-X(SHEV3), SKHEV2-H(SHEV3), and SKHEV2-X(SHEV3) (Figure 1). In each of these four chimeras, the genomic backbone is HEV-1 (Sar-55 strain) which only infects humans, while the entire ORF1 or its functional domain regions were swapped with the corresponding regions from HEV-3 (Meng strain) which infects both humans and pigs.43 Therefore, if the ORF1 regions are determinant(s) of HEV host range, the chimeric virus(es) are expected to acquire the ability to infect pigs. Also, to investigate if the human ribosomal protein sequence S17 insertion in the ORF1 HVR can expand HEV host tropism, we also constructed a recombinant clone SKHEV2(S17) by insertion of the S17 sequence in the HVR of the HEV-1 genomic backbone (Figure 1).
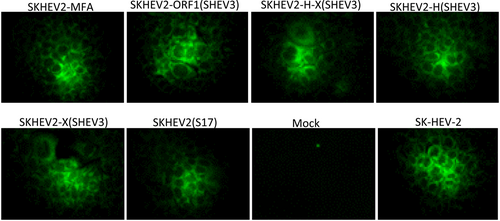
The full-length capped genomic RNA transcripts from each of the intergenotypic chimeric clones, the recombinant clone SKHEV2(S17), as well as the modified parental clone SKHEV2-MFA were transfected into Huh7-S10-3 human liver cells to determine the replication competency of the constructs in vitro. Approximately 6 days posttransfection, HEV-specific proteins were detected in transfected cells by using a HEV-specific hyperimmune chimpanzee antiserum. As shown in Figure 2, similar to the parental clone SK-HEV-2, the modified clone SKHEV2-MFA also showed positive IFA signals. Similar to the SKHEV2-MFA, all the four intergenotypic chimeric clones (SKHEV2-ORF1(SHEV3), SKHEV2-H-X(SHEV3), SKHEV2-H(SHEV3), and SKHEV2-X(SHEV3)) as well as the S17 recombinant clone SKHEV2(S17) produced HEV-specific IFA signals, while the mock-transfected negative control cells remained negative (Figure 2). The results indicated that the insertions of three enzyme sites did not abolish the infectivity of the HEV-1 backbone clone, and that all the four intergenotypic chimeric clones and the S17 recombinant clone SKHEV2(S17) are replication competent in human liver cells in vitro.
3.2 The intergenotypic chimeric viruses failed to infect pigs
To investigate if the intergenotypic chimeras acquire the ability to infect pigs, the RNA transcripts from each of the chimeras were intrahepatically inoculated into the liver of pigs (Table 1). Previously we had successfully used the intrahepatic inoculation of capped RNA transcripts into the liver of pigs to test for the infectivity and pathogenicity of chimeric or recombinant HEVs.37, 43 The fecal and serum samples from each pig were collected before inoculation and weekly thereafter, and tested for the presence of HEV RNAs. As shown in Table 2, as expected, the HEV-3 parental clone SHEV3-inoculated pigs (ID no. 9, 10) in the positive control group developed transient viremia starting from 3 wpi, and the third pig (ID no. 16) in the positive control group also became viremic starting from 4 wpi. Similar to viremia, the fecal virus shedding in the positive control group started from 3 wpi in two pigs (ID no. 9, 10), and 4 wpi in the third pig (ID no. 16). The viremia and fecal virus shedding became undetectable at 6 and 7 wpi, respectively. No viral RNA was detected in samples of liver and bile collected at necropsy at 10 wpi.
| No. of positive serum/fecal samples at wpi (liver/bile at necropsy) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | Pig no. | 0 wpi | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Liver/bile |
| PBS | 3 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
| SKHEV2-MFA | 3 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
| SHEV3 | 3 | 0/0 | 0/0 | 0/0 | 2/2 | 3/3 | 2/3 | 0/2 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
| SKHEV2(S17) | 3 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
| SKHEV2-ORF1(SHEV3) | 3 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
| SKHEV2-H-X(SHEV3) | 3 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
| SKHEV2-H(SHEV3) | 3 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
| SKHEV2-X(SHEV3) | 3 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
- Abbreviations: HEV, hepatitis E virus; PBS, phosphate-buffered saline; wpi, week postinoculation.
The IgG anti-HEV antibodies in sera were detected in all three pigs from the positive control group starting from 5 wpi (Figure 3). As expected, the negative control groups, which consist of the PBS-inoculated pigs as well as the HEV-1 parental backbone SKHEV2-MFA-inoculated group, remained negative for HEV viremia, fecal virus shedding, or seroconversion to anti-HEV antibody throughout the study (Table 2; Figure 3). None of the pigs inoculated with the intergenotypic chimeras had detectable HEV RNA in serum or fecal samples throughout the study or in liver and bile samples collected at necropsy (Table 2). Similarly, there was no seroconversion to IgG anti-HEV in any of the pigs inoculated with the intergenotypic chimeras (Figure 3). The results suggested that the HEV-1-based intergenotypic chimeras containing the swapped ORF1 regions from the zoonotic HEV-3 did not gain the ability to infect pigs.
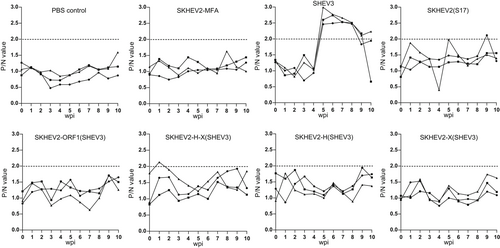
3.3 The recombinant virus SKHEV2(S17) with the S17 insertion did not acquire the ability to infect pigs
Under in vitro conditions, it has been reported that insertion of S17 in the HEV HVR expanded the host range allowing the recombinant virus to infect cells from different animal origins.39 Therefore, to investigate if the recombinant virus SKHEV2(S17) with an S17 insertion in the HVR of HEV will gain the ability to replicate in vivo, the RNA transcripts of the recombinant clone SKHEV2(S17) were intrahepatically inoculated into the liver of pigs (Table 1). Similar to the results from the intergenotypic chimeras, the inoculated pigs did not develop HEV viremia or fecal virus shedding and did not seroconvert to HEV antibody (Table 2; Figure 3). Therefore, the results indicated that the S17 recombinant virus SKHEV2(S17) did not gain an expanded infection capability in pigs.
4 DISCUSSION
Zoonotic HEV infections, especially by HEV-3 and HEV-4, are associated with foodborne hepatitis, chronic hepatitis as well as neurological diseases.9, 10, 26, 47 The HEV has an extensive genetic diversity and host range.15, 16 The HEV-3 and HEV-4 are zoonotic and infect across species barriers. Particularly for HEV-3, a large number of animal reservoirs, such as pigs, rabbits, mongooses, and deer have been identified.16 In contrast, the HEV-1 and HEV-2 are restricted to humans, and not known to infect other animal species.
The viral and host factors responsible for HEV cross-species infection remains unknown. Since the HEV ORF2 gene encodes the only structural protein (capsid), and binds to cellular receptor, and thus the ORF2 gene was presumed to determine the HEV host range and tropism.36, 48, 49 However, this presumption was challenged by two of our previous studies.37, 38 We showed that intergenotypic chimeric viruses containing the ORF2 and/or ORF3 gene of HEV-3 and HEV-4 strains in the genomic backbone of HEV-1 failed to infect pigs, suggesting that the viral genetic element responsible for HEV cross-species infection apparently does not reside in the ORF2/ORF3. Additionally, other research showed that the broad host range of zoonotic HEV-3 and HEV-4 is associated with extensive co-evolution of viral genome.50, 51 For example, amino acid at position 1252 in ORF1 is recognized as a HEV-3 host-specific motif.50 The available evidence suggests that the ORF1 nonstructural gene may be involved in the HEV host tropism. Therefore, the main objective of this present study was to determine if chimeric viruses in the genomic backbone of the human-exclusive HEV-1 bearing the ORF1 or its functional domain regions of zoonotic HEV-3 will acquire the ability to cross species barrier and infect pigs.
To achieve our main objective, intergenotypic chimeric viruses were constructed by utilizing the unique biological property of HEV-1 and HEV-3. The backbone virus is based on HEV-1 (strain Sar-55; pSK-HEV-2) that infects only humans but does not infect pigs.42 The donor sequences for gene swaps in the HEV-1-based chimeric viruses include the ORF1 and its functional domain regions from the zoonotic HEV-3 (Meng strain) that infects both humans and pigs. Therefore, if the ORF1 or its functional domain region is involved in HEV cross-species infection, then the chimeric viruses containing the genetic element(s) from HEV-3 theoretically should acquire infectivity in pigs. To bypass the use of in vitro cell culture system to propagate HEV which is insufficient, we utilized the in vivo transfection procedure by direct intrahepatic inoculation of capped full-length HEV RNA transcripts directly into the liver of pigs to test for the infectivity of the chimeras.37, 43 Four chimeric viruses: SKHEV2-ORF1(SHEV3), SKHEV2-H-X(SHEV3), SKHEV2-H(SHEV3), and SKHEV2-X(SHEV3) were constructed. We demonstrated that capped RNA transcripts from all chimeric viruses were replication competent in Huh7 liver cells. This suggests that swapping the ORF1, and its functional domains (HVR and X) within ORF1 between HEV-1 and HEV-3 did not affect the replication competency of the genotype 1 HEV-based intergenotypic chimeric clones. However, none of the intergenotypic chimeras was able to infect pigs as evidenced by the lack of viremia, fecal virus shedding or seroconversion to HEV antibody in any of the inoculated pigs, even though all pigs intrahepatically inoculated with the parental HEV-3 clone developed viremia and fecal virus shedding, and also seroconverted to IgG anti-HEV. The use of a high sensitivity nested RT-PCR assay minimized the possibility of missing a positive detection in samples with low viral RNA loads. The results from this study suggest that the ORF1 gene and its HVR and X domains, are not solely involved in determining the HEV host tropism.
It has been reported that human ribosomal protein sequences, such as S17 and S19 are naturally inserted in the genome of human HEV-3 strains.39, 40, 52 Importantly, it was shown that insertion of S17 in the HVR of HEV-1 expanded the host range of the recombinant virus enabling it to infect cells from different animal species including pig and deer in vitro.39, 53 We also demonstrated that the S17 sequence contains a nuclear localization signal, and that insertion of S17 sequence in the HVR of HEV ORF1 bestowed novel nuclear/nucleolar trafficking capabilities to the ORF1 protein of a human HEV-3 strain Kernow P6.40, 54 Therefore, in this present study we also aimed to determine if the recombinant HEV-1 containing the S17 sequence will acquire the ability to expand the host range and infect pigs. Again, we showed in this study that the recombinant virus SKHEV2(S17) which contains S17 failed to infect pigs, as the inoculated pigs had no evidence (viremia, fecal virus shedding and seroconversion) of virus replication. This result suggested that the S17, which can expand HEV tropism in vitro, is not a determinant factor for HEV cross-species infection in vivo.
The results from this study further highlighted the complexity regarding the mechanism of HEV cross-species infection. There are several possible explanations and limitations in this study. First, as other studies have revealed that the expanded host range of HEV was associated with extensive co-evolution of whole virus genome.50, 51 It is possible that critical amino acid mutations, which may occur in both viral ORF2 structural protein and ORF1 nonstructural protein, cooperatively play a role in determining HEV host tropism. Second, the molecular structure of HEV is still not fully understood.14 Novel coding genes, such as ORF4 in HEV-1, which is located within ORF1, was identified recently.35 Therefore, other unidentified gene coding structural or nonstructural proteins may also play a role in determining HEV host tropism. Third, technically, the HEV-1 backbone infectious clone pSK-HEV-2 used in this study and our previous two studies has a low virus replication efficiency both in vitro and in vivo.37, 38, 42 This may affect the robust infection and replication of chimeric viruses in pigs, especially if the chimeric viruses replicate at a low level. Fourth, in addition to the viral genetic elements, host factors either alone or in association with viral genetic element(s) may also be important in mediating HEV cross-species infection. Nevertheless, the results from this study did reveal that the HEV genome has a relatively high flexibility, since insertions of restriction enzyme sites or long foreign genes sequence, and exchange of heterologous gene from different genotypes did not apparently abolish the replication competency of the backbone virus in vitro. The results from this study will help us further understand the underlying mechanism of cross-species infection by HEV.
ACKNOWLEDGMENTS
The authors wish to thank Alex Bishop, Delaney Conrad, Dr. Alessandra M.M.G. Castro and Laboratory Animal Resources staff at Iowa State University for their help in the animal study. We thank Drs. Suzanne U. Emerson and Robert H. Purcell at the National Institutes of Health (Bethesda, MD) for kindly providing us with the HEV-1 infectious clone pSK-HEV-2, Huh7-S10-3 cells, and anti-HEV hyperimmune chimpanzee 1313 antiserum. This study is supported by grants from the National Institutes of Health (R01 AI074667, and R01 AI050611). Danielle M. Yugo is supported by a Ruth L. Kirschstein National Research Service Award Institutional Research Training Grant (T32OD010430). Tanja Opriessnig is supported by the Biotechnology and Biological Sciences Research Council (BBSRC) through the Roslin Institute Strategic Program “Control of Infectious Diseases” (BBS/E/D/20002173 and BBS/E/D/20002174).
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
DT and DMY contributed to the design, implementation and data analysis of the study, and wrote the manuscript. XJM contributed to the design of the study, and wrote the manuscript. SPK contributed to the study design. DT, DMY, CLH, TO, AKK, JB, and PGH contributed to the execution of the animal experiment. All authors reviewed and commented on the manuscript, and approved for publication.