Acute gastroenteritis outbreak associated to norovirus GI.9 in a Portuguese army base
Abstract
Gastroenteritis is considered a major illness within the military settings being caused by foodborne enteric pathogens that are particularly easily spread in the crowded conditions of military camps. Gastroenteritis outbreaks caused by norovirus usually affect a great number of soldiers due to the low infectious dose, copious viral shedding, and environmental stability. The present study describes the investigation of an outbreak of acute gastroenteritis that occurred in April 2015 in a Portuguese army base, focusing on the study of the epidemiological curve, symptoms experienced by the affected soldiers, and results of food, water, and stool microbiological analysis. From a total of 938 military personnel stationed on the base 46 soldiers developed acute gastroenteritis. Stool analysis of seven cases showed to be positive for norovirus GI.9 that was the probable cause of the outbreak. This report shows that genogroup I norovirus can also cause considerable morbidity in healthy young soldiers, affecting the operational effectiveness on the military forces. J. Med. Virol. 89:922–925, 2017. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Gastroenteritis is considered a major illness within the military settings being caused by foodborne enteric pathogens that are particularly easily spread in the crowded conditions of military camps [AFHSC, 2013a]. Many efforts have been employed to reduce food-related illness with special focus on the employment of protective measures, education of good food handling practices, and the implementation of early-warning systems. However, gastroenteritis remains a central concern in morbidity among military forces and of all the enteric pathogens, noroviruses were found to be the most common cause of gastroenteritis in both U.S. and U.K. troops [Bailey et al., 2008; AFHSC, 2013a,2013b]. Gastroenteritis outbreaks caused by norovirus usually affect a great number of people due to the low infectious dose, copious viral shedding, and environmental stability [Hall, 2012]. The high morbidity rendering a high number of military personnel non-effective, diminishing operational effectiveness, and impacting on force readiness [Thornton et al., 2005; Bailey et al., 2008]. The present study describes the investigation of an outbreak of acute gastroenteritis that occurred in April 2015 in a Portuguese army base, focusing on the study of the epidemiological curve, symptoms experienced by the affected soldiers, and results of food, water, and stool microbiological analysis.
MATERIALS AND METHODS
Epidemiological Investigation
On April 23, 2015, the Portuguese Army Laboratory of Bromatology and Biological Defence (LBBD) was notified of an acute gastroenteritis outbreak in a military base that started 3 days before (20 April). Stool specimens, food items, and water were collected during the outbreak and a retrospective investigation was initiated. An anonymous questionnaire was conducted to gather epidemiological and clinical data asking soldiers to list clinical symptoms and disease onset. A case was defined as a member of the military unit staff who presented, in the period between April 20 and 24, 2015, at least one measurable symptom of gastroenteritis (three or more liquid stool in 24 hr, a vomiting episode or temperature ≥37°C), have eaten at the base canteens, and have attended the base medical center. The base had a total staff of 983 military personnel divided in several units, each with its own canteen (A, B, C) and cuisine, where food meals were prepared and served. Food items used in the three canteens had the same origin, but were prepared by different cooking teams. Soldiers were not allowed to eat in canteens from other companies.
As per standard procedures in the military bases, samples of served food are routinely stored in the refrigerator during a 48-hr period for future testing, if required. As notification to LBBD occurred 3 days after the first symptomatic case, food samples were no longer available for microbiological analysis. In an effort to investigate the origin of the outbreak and despite having passed 3 days from the index case (April 20), the LBBD collected samples from the dinner served April 21 in canteen A and the meals served the April 23 and 24 of canteen B. No food was sampled from canteen C because no cases were reported there. Tap water samples were collected from canteen A and B on the April 24. Stools from seven symptomatic soldiers (Four eating in canteen A and three in canteen B) were also collected: six individual specimens on April 23 and one on April 24.
Laboratory Investigation
Food samples were tested for bacteria namely, Salmonella spp. (Vidas® Salmonella [SLM], bioMerieux, Marcy-l'Etoile, France), Enterobacteriaceae (ISO 21528-2:2004), Escherichia coli (ISO 16649-2:2001), Bacillus cereus (ISO 7932:1987), Clostridium perfringens (ISO 7937:1997), Campylobacter spp. (Vidas® Campylobacter (CAM), bioMerieux), Staphylococcus aureus (ISO 6888-1:1999), and Listeria monocytogenes (Vidas® L. monocytogenes II, bioMerieux). Water was processed according to standard membrane filter procedures for enumeration of culturable microorganisms (ISO 6222:1999), E. coli (ISO 9308-1:2000), coliform bacteria (ISO 9308-1:2000), Pseudomonas aeruginosa (ISO 16266:2006), faecal Streptococci (ISO 7899/2:1984), and detection/enumeration of spores of sulfite-reducing anaerobes (Clostridia) (ISO 6461-2:1986). Neither food nor water was tested for the presence of enteric viruses due to the exhaustion of samples in bacteriological analysis.
Stools were screened for common enteropathogenic bacteria, namely Shigella, Salmonella, Campylobacter, Vibrio, B. cereus using standard coproculture methods. Counting of C. perfringens in stools was also performed. Stools were also searched for enteropathogenic viruses, namely norovirus, astrovirus, rotavirus, adenovirus, and sapovirus using a multiplex Real-Time PCR assay (FTD Viral Gastroenteritis, Fast Track Diagnostics, Luxembourg) as previously described [Lopes-João et al., 2015]. Stool samples that showed to be positive for norovirus were submitted to conventional RT-PCR assays, and subsequently sequenced and typed. Briefly, after two conventional RT-PCR targeting a 747 bp fragment of ORF1 gene that encodes a region of the RNA-dependent RNA polymerase (RdRp) and a 268 bp fragment of the ORF2 gene that encodes a region of the capsid protein [Vinjé et al., 2004; Fonager et al., 2013], amplified products of the expected sizes were sequenced (Sequencer Analyser ABI-Prism 3130 xl, PE Applied Biosystems, LLC, Foster City, CA). Genotypes were assigned using a public automated genotyping tool (http://www.rivm.nl/en/Topics/N/NoroNet/Databases/Sequence_typing_tool) from Noronet platform developed by the National Institute for Public Health and the Environment (RIVM) [Kroneman et al., 2011].
RESULTS AND DISCUSSION
From the total of 938 military personnel that were stationed on the base 46 soldiers met the case definition, 26 of which have eaten at canteen A and 20 at canteen B.
The first case of this outbreak occurred on April 20 in a soldier that ate on canteen B (Fig. 1). On this canteen, the majority of cases occurred on the April 22 (nine cases) being the last cases reported on the April 24 (two cases). Regarding canteen A, the first case occurred in April 21, the majority of cases occurred on April 23 (16 cases), and the last cases were reported on April 24 (four cases). The bacteriological analysis of the food served at canteen B indicated good microbiological quality but dinner served at canteen A (baked pork meat with saffron sauce and rice) presented an enumeration of C. perfringens of 1.8 × 106 CFU/g, a level above the acceptable microbiological limits and considered potentially hazardous. However, the batch of frozen raw pork and saffron used to prepare this dinner presented microbiological values within acceptability, with no C. perfringens detected, suggesting that the high C. perfringens levels founded in baked pork meat was likely caused by inadequate food handling practices.

The bacteriological analysis of the tap water from canteens A and B, that was only sampled at the end of the outbreak (on April 24), was of acceptable quality.
However, we found that an additional water source for the canteens that used groundwater well that was positioned inside the base had been consumed until April 23 and showed an interesting connection to the outbreak. Canteen B, that registered the first case of the outbreak, distanced approximately 1 km from this groundwater well, while canteen A (that registered cases 24 hr later) distanced approximately 2 km, and canteen C (with no cases) approximately 3 km. We could not ascertain whether distance-factor was involved in the transport of a waterborne enteropathogenic agent from the point source (the groundwater well) with the consequent concentration decrease of the contaminated water along the length of the pipes. However, the time difference in the appearance of first case in canteens B versus A, as well as the peaks and last cases, could be justified by the closest proximity of the well to canteen B, compared to canteen A. This hypothesis is supported by the absence of cases in the furthest canteen C (3 km). Unfortunately, samples from the groundwater well were not provided for analysis.
Among the seven stool specimens that were available for analysis only one presented high C. perfringens count (2.6 × 107 CFU/g) that could be considered of clinical significance (>106 CFU/g). This stool was from a soldier that had dinner at canteen A, where the pork meat with high C. perfringens counts was served. Since high counts were only found in the stools of one soldier, C. perfringens can be ruled out as the cause of the outbreak. Concerning enteric viruses detection, all seven stool specimens showed to be positive for norovirus GI. No other enteric viruses were found in these stools. Subsequent genotype characterization in both partial ORF1 and ORF2 regions identified all isolates as GI.9 (Figs. Fig. 2 and 3), with 100% nucleotide sequence homology, suggesting that this GI.9 strain was the most probable cause of the gastroenteritis cases of both canteens and that all cases had the same origin. The contiguous genomic region sequence assembled using the amplified nucleotide sequences of the polymerase and capsid regions has been deposited in GenBank under accession number KX458103.
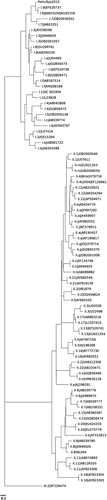
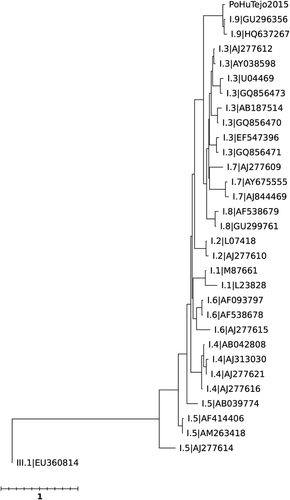
This investigation was intricate because the outbreak affected soldiers that ate at different canteens, each with its own team of food handlers. Moreover, the exhaustion of samples on bacteriological analysis (traditionally the first pathogens to be searched) did not left any food or water available for virological analysis that could have allowed a definite conclusion on the origin. Poor hygienic manipulation of food was suspected in canteen A based on the presence of C. perfringens in the meal and its absence in the raw materials used to prepare it. However, this outbreak cannot be ascribed to poor hygiene in food handling in canteen A, since each canteen has its own staff and the bacteriological analysis of the food served at canteen B indicated good microbiological quality. It would be tempting to attribute the norovirus GI.9 transmission to an infected food handler, but this is highly unlikely since different cooking teams prepared the food (one in each unit/canteen) and soldiers were not allowed to eat in canteens from other companies.
Based on all the data, it is tempting to speculate that this GI.9 norovirus outbreak might have been waterborne and that the groundwater that was supplied to the canteens could be the point source of the norovirus contamination. Unfortunately, this hypothesis cannot be confirmed, since the analyzed water was only sampled at the end of the outbreak, after the alert, and adjustment of disinfection measures. This report shows the importance of continuous epidemiological surveillance and swift intervention in military settings, as well as the impact of norovirus GI.9 in the morbidity of military personnel.
ACKNOWLEDGMENT
None.




