Epidemiology of rotavirus A diarrhea in Chókwè, Southern Mozambique, from February to September, 2011
Abstract
Acute diarrhea disease caused by Rotaviruses A (RVA) is still the leading cause of morbidity and mortality in children ≤5 years old in developing countries. An exploratory cross-sectional study was conducted between February and September, 2011 to determine the proportion of acute diarrhea caused by RVA. A total of 254 stool specimens were collected from children ≤5 years old with acute diarrhea, including outpatients (222 children) and inpatients (32 children), in three local health centers in Chókwè District, Gaza Province, South of Mozambique. RVA antigens were detected using enzyme immunoassay (EIA); the RVA G (VP7) and P (VP4) genotypes were determined by RT-PCR or analysis sequencing. Sixty (24%) out of 254 fecal specimens were positive for RVA by EIA; being 58 (97%) from children ≤2 years of age. RVA prevalence peaks in June and July (coldest and drier months) and the G[P] binary combination observed were G12P[8] (57%); G1P[8] (9%); G12P[6] (6%); and 2% for each of the following genotypes: G1P[6], G2P[6] G4P[6], and G9P[8]. Non-Typeable (NT) G and/or P genotypes were observed as follows: G12P [NT] (6%); G1P [NT], G3P[NT] and GNTP[NT] (4%). Considering the different GP combinations, G12 represented 67% of the genotypes. This is the first data showing the diversity of RVA genotypes in Mozambique highlighting the epidemiological importance of these viruses in acute diarrhea cases in children ≤2 years old. In addition, these findings will provide a baseline data before the introduction of the RVA monovalent (Rotarix®) vaccine in the National Immunization Program in September 2015. J. Med. Virol. 88:1751–1758, 2016. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Rotaviruses A (RVA) are the most important cause of severe acute diarrhea in children ≤5 years old worldwide and it is associated with 28% of deaths in children of this age group, particularly in Asia and Africa [UNICEF, 2013]. In Mozambique, a previous study reported that acute diarrhea was the third cause of childhood deaths and RVA was the leading cause of death during the first 2 years of life [Kotloff et al., 2013].
Rotavirus is a genus of Reoviridae family. Its genome consists of 11 segments of double stranded RNA (dsRNA) surrounded by a triple-layered capsid, without an envelope. Among the proteins encoded by these genes, six are structural: VP1-VP4, VP6, and VP7; and six non-structural: NSP1- 6 [Estes and Greenberg, 2013].
So far, 27 G and 37 P RVA genotypes infecting different species have been described and the genotypes distribution varies across different continents. Six binary combinations: G1P[8], G2P[4], G3P[8], G4P[8], G9P[8], and recently G12P[8] that emerged in different continents, are considered the most prevalent human RVA genotypes [Banyai et al., 2007; Rahman et al., 2007; Matthijnssens et al., 2008; Banyai et al., 2012; Matthijnssens and Van Ranst, 2012; Ndze et al., 2013; Trojnar et al., 2013; Bucardo et al., 2015; Delogu et al., 2015]. In Africa, the first RVA G12 genotype was reported in South Africa and subsequently in other African countries [Cunliffe et al., 2009; Page et al., 2009; Banyai et al., 2012; Seheri et al., 2014].
The World Health Organization has recommended the implementation of a country-wide surveillance for RVA prevalence and circulating genotypes before and after the introduction of the RVA vaccine in their National Immunization Program (NIP) [WHO, 2008]. The African RVA surveillance network was established in 2006, when some countries joined it and data concerning RVA burden became available [Mwenda et al., 2010; Seheri et al., 2014].
So far, twenty-nine African countries have introduced the RVA vaccine in their NIP (http://sites.path.org/rotavirusvaccine/files/2015/12/PATH-Country-Introduction-Table-EN-2016.01.01.pdf). The preliminary data from these countries showed that RVA vaccines were safe and effective in protecting children from severe diarrhea caused by RVA. However, the impact of regionally prevalent and mixed genotypes on vaccine effectiveness in Africa is not yet clear, and needs to be monitored [Madhi et al., 2010; Cunliffe et al., 2012; Groome et al., 2014; Page et al., 2014; Bar-Zeev et al., 2015].
In Mozambique, there is no available data describing RVA epidemiology and surveillance studies are important to understand its broader impact on circulating strains to reinforce the measures for diarrhea disease control currently in place. In September 2015, the monovalent vaccine (RV1; Rotarix™ GlaxoSmithKline (GSK) Biologicals, Rixensart, Belgium) was introduced in Mozambique NIP. Therefore, on aiming to contribute with information concerning RVA epidemiology, we conducted the present study to determine RVA prevalence and genotypes circulating in Chókwè District, Southern part of the country, to represent the first baseline preceding the introduction of the RV1 in Mozambique.
MATERIALS AND METHODS
Study Population
This cross-sectional study was conducted in Chókwè District, Gaza Province, Southern Mozambique, from February to September, 2011, where the warmer and rainy seasons occur from November to April; and the coldest and drier seasons occur from May to October. A total of 254 fecal samples were collected from children ≤5 years of age with acute diarrhea, being 222 (87%) from outpatients and 32 (13%) from inpatients. All children received medical assistance at Chókwè Rural Hospital Centre or at Carmelo Hospital. Diarrhea was defined as three or more loose or watery stools occurred within a 24 hr period. Other causes were also enrolled in the study. Children with other symptoms than those, such as evacuation accompanied by loose or watery stools presenting traces of visible blood were not included. Acute occurrence was defined as an episode of diarrhea lasting less than 14 days and onsets lasting 14 days or more was considered persistent [WHO, 2005]. A written informed consent before enrolment was obtained from parents or legal guardians of each participant. This study received the Ethical approval from the Mozambique National Bioethics Committee (IRB00002657, reference No: 384/CNBS/10).
Fecal Specimen's Collection, Rotaviruses A Detection and Genotype Characterization
Fecal specimens were collected from childreńs diapers, transferred to two polystyrene tubes, kept refrigerated in cooler boxes, and then transported to the laboratory of the Chókwè Health Research and Training Centre, within 1 hr of collection.
For water diarrhea, a portion of the diaper was dissolved in saline solution (0.9%). One of the tubes was stored at 4°C before tested by enzyme immunoassay (EIA) for RVA and the second tube was stored at −20°C for later molecular characterization. The EIA (ProSpecT™ Rotavirus Kit, Oxoid, Ltd, United Kingdom) was carried out as recommended by the manufacturer. The positive samples by EIA were later shipped in dry ice to the Laboratory of Comparative and Environmental Virology, Oswaldo Cruz Institute, Fiocruz, Brazil, for RVA molecular characterization.
Nucleic acids were extracted by the glass powder method [Boom et al., 1990], including modifications [Leite et al., 1996]. The dsRNA was reverse transcribed and amplified by PCR using a pair of consensus primers corresponding a conserved nucleotide sequence of the VP7 gene: Beg9-End9 (1064 bp) [Gouvea et al., 1990] or 9con1-9con2 (922 bp) [Das et al., 1994]; VP4 gene: 4Con2-4Con3 (686 bp) [Gentsch et al., 1992] and SuperScript™ III One-Step RT-PCR System with Platinum® Taq DNA Polymerase kit (Invitrogen®, Carlsbad, CA) following the manufacturer's instructions. The RVA were genotyped by seminested-PCR using the primers and protocols described previously for VP7 [Gouvea et al., 1990; Das et al., 1994] and VP4 [Gentsch et al., 1992] genes.
Nucleotide Sequencing and Phylogenetic Analysis
Specimens that could not be genotyped by seminested-PCR were subjected to nucleotide sequencing using individually the consensus G and P primers. First round products of RT-PCR (consensus amplicons) were purified with QIAquick™ PCR Purification Kit (QIAGEN™, Valencia, CA), according to the manufacturer's instructions and sequenced using the Big Dye Cycle Sequencing reaction kit™ (Applied Biosystems, Foster City, CA) and ABI Prism 3730 Genetic Analyzer™ (Applied Biosystems). The nucleotide sequences obtained were aligned and edited using the Bio-Edit Sequence Alignment Editor (version 6.0.5.2) then compared with corresponding sequences of selected rotavirus strains available in the GenBank database.
The phylogenetic analysis was performed using Mega 4.0 [Tamura et al., 2007] and the neighbor-joining method with distances calculated by the Kimura-2 parameter method. Statistical support was assessed by bootstrapping with 2,000 replicates.
The GenBank accession numbers of the sequenced samples were as follows: (i) for VP7 gene: KP222808−KP222852; (ii) for VP4 (VP8*) gene: KP222853−KP222878.
RESULTS
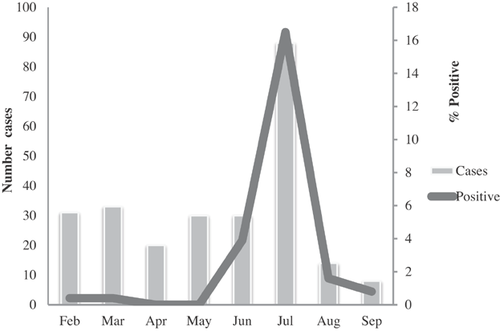
Of 254 fecal samples obtained from children ≤5 years old with acute diarrhea enrolled in the study, 60 (24%) were positive for RVA by EIA; being 58 (97%) from children ≤2 years of age (Table I). RVA prevalence peaked in June and July (coldest and drier months) (Fig. 1).
| Age (months) | RVA Positive/N (%) |
|---|---|
| 0–6 | 13/72 (18) |
| 6–12 | 30/79 (38) |
| 12–24 | 15/88 (17) |
| >24 | 2/15 (13) |
| Total | 60/254 (24) |
The G[P] binary combination detected was: G12P[8] (57%); G1P[8] (9%); G12P[6] (6%); and 2% for each of the following genotypes: G1P[6], G2P[6] G4P[6] and G9P[8]. Non-Typeable (NT) G and/or P genotypes were observed as follows: G12P[NT] (6%); G1P[NT], G3P[NT] and GNTP[NT] (4%) (Fig. 3). The samples in which GP genotype was not obtained by semi nested-PCR were submitted to sequencing (n = 21). However, some of them remained NT. Considering the different GP combinations, G12 represented 67% of the genotypes (Table II). Seven samples could not be genotyped due to the amount availability of fecal material.
| Genotypes | G1 | G2 | G3 | G4 | G9 | G12 | GNT | Total (%) |
|---|---|---|---|---|---|---|---|---|
| P[8] | 5 | 0 | 0 | 0 | 2 | 30 | 1 | 38 (71.7) |
| P[6] | 1 | 1 | 0 | 1 | 0 | 3 | 0 | 6 (11.3) |
| P[4] | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 (1,9) |
| P[NT] | 2 | 0 | 1 | 0 | 0 | 3 | 2 | 8 (15,1) |
| Total (%) | 8 (15) | 1 (2) | 1 (2) | 1 (2) | 2 (4) | 36 (68) | 4 (8) | 53 (100) |
- NT, non-type able.
Phylogenetic analysis based on VP7 and VP4 (VP8*) nucleotide sequences showed a great diversity of Mozambican strains (Fig. 2) in 2011. Analysis of the gene encoding the VP7 protein (Fig. 2A) showed that G12 strains were classified into two different groups inside the G12-III lineage: (i) one group clustering together with prototype strains detected in several countries such as Thailand, Uganda, South Africa, and Zimbabwe; (ii) one group clustering together with one G12P[6] strain detected in India in 2009 and one G12P[8] strain detected in Mauritius in 2011. The G2 study strain (MOZ21178) belongs to the G2-I lineage. The two G9 strains (MOZ21155 and MOZ21162) clustered inside the G9-III, but into two different groups: MOZ21155 strain clustered together with one strain detected in Germany in 2008 and one strain detected in Ethiopia in 2012; the MOZ21162 strain grouped together with strains detected in Bangladesh, Bhutan, India, and South Africa between 2008 and 2010. The G4P[6] strain (MOZ21205) clustered with human strains detected in 2009 in India (KOL-78-09), in 2013 in China (R1954), and Thailand (CU-B1738). All Mozambican G1 strains clustering together with G1-I lineage of human prototype strains were detected worldwide.


Concerning the phylogenetic analysis of the gene encoding the VP8* portion of VP4 protein (Fig. 2B) of Mozambican strains, three P[6] strains grouped inside the P[6]-I lineage, but into two different clusters: (i) MOZ21205 grouped with three porcine prototype strains (one detected in 2004 in Italy, one detected in Japan in 2006, and one detected in China 2008 and one human prototype strain detected in China in 2004; (ii) the two other P[6] strains (MOZ21168, MOZ21178) clustered together with human prototype strains were detected in Bangladesh, Burkina Faso, Germany, India, South Africa, and USA. The P[4] strains were closely related with human prototype strains detected in Europe and North America. P[8] sequences were clustered into two different lineages: MOZ21162 inside the P[8]-4 lineage and the others into the P[8]-3 lineage.
DISCUSSION
The present study shows a high prevalence and genotyping diversity of RVA in children ≤5 years of age living in Chókwè District, Southern Mozambique, and corroborates findings from a previous study conducted in the country showing the importance of RVA in acute diarrhea disease [Rappelli et al., 2005]. In our study, RVA infections were detected more frequently in children between 7 and 12 months of age, similar to reports from other areas of Southern Africa [Potgieter et al., 2010; Waggie et al., 2010].
A peak of RVA infection in Chókwè was observed between June and July, which are the coldest and drier months. This pattern appears to be consistent with that reported in Mozambique [Kotloff et al., 2013] and other African countries [Armah et al., 1994; Patel et al., 2012]. One of the limitations of the present report was the study period, comprising the months from February to September. However, due to logistic constraints we decided to conduct the study in this period, corresponding the driest season of the year.
In this study, RVA genotype G12P[8] was the prevalent genotype, corroborating previous reports conducted in Asian, European, American, and African continents, demonstrating the epidemiological relevance of this genotype worldwide [Cunliffe et al., 2009; Page et al., 2009; Potgieter et al., 2010; Jere et al., 2011; Page et al., 2014; Seheri et al., 2014; Bucardo et al., 2015; Luchs et al., 2015]. The high proportion of genotype G12 in African populations is believed to be associated with the emergence of genotype P[6] [Steele and Ivanoff, 2003; Page et al., 2009; Seheri et al., 2014]. RVA genotypes G12P[8] and G12P[6] were previously reported in some neighboring African countries such as Malawi and South Africa [Page et al., 2009, 2014; Kiulia et al., 2014]. Otherwise, the G12P[6] Mozambican strains, besides the G12P[8] strain MOZ21153, showed to be more related with strains detected in India and Mauritius in 2009 and 2012, respectively, forming a second cluster inside the G12-III lineage. The remaining G12P[8] strains grouped inside the G12-III together with human prototype strains detected in different continents in the last decade corroborating previous studies showing G12-III circulation in the African continent [Page et al., 2009; Komoto et al., 2014; Ndze et al., 2014].
The two G9P[8] clustered into the G9-III lineage, corroborating different studies showing the circulation of this lineage into different African countries [Esona et al., 2013; Komoto et al., 2014; Nyaga et al., 2014]. Otherwise, sequences grouped into two different clusters inside G9-III: MOZ21162 grouped with human strains detected in Bangladesh, Bhutan, India, South Africa, and Zimbabwe between 2008 and 2011 and into the other cluster, MOZ21155 grouped together with the strain GER176-08 detected in Germany in 2008, recovered from diarrheic feces of a 67-year-old man and the MRC-DPRU842 strain, detected in Ethiopia in 2012, recovered from diarrheic feces of a 16-month-old girl.
Genotype G4 is commonly detected in porcine associated with P[6] worldwide [Papp et al., 2013; Heylen et al., 2014; Martinez et al., 2014; Zhou et al., 2015]. Otherwise, due to the close relationship between human and pigs it is not rare to detect genotypes originally related to porcine in humans, as they seem to share the same origin in case of genotypes belonging to the Wa-like genogroup. Corroborating previous findings, the G4 Mozambican strain showed close genetic relation with other human porcine-like strains detected in 2009 in India (KOL-78-09) and in 2013 in China (R1954) and Thailand (CU-B1738), reinforcing the hypothesis that human and porcine RVA may have a common origin. Further analysis will be carried out to this specific strain (characterization of the 11 RVA genes).
The analysis of the G1 nucleotide sequences showed the exclusive circulation of the G1-I lineage into the Mozambican strains. They clustered separately, into two groups: (i) the first one with G1P[8] strains MOZ21135 and MOZ21123 grouping together with other G1-I strains detected worldwide in the last decades; (ii) the second with the two G1P[X] strains MOZ21142 and MOZ21120 and the G1P[6] strain MOZ21191 clustering with two strains detected in Bhutan in 2010, one strain detected in Brazil in 2012 and one strain detected in Ethiopia with unavailable date of collection. The present results corroborate previous studies reporting the circulation of the G1-I lineage worldwide [Ndze et al., 2014; Abdel-Moneim et al., 2015; Silva et al., 2015].
Analysis on VP8* portion of the VP4 protein revealed the co-circulation of three genotypes (P[8], P[4], and P[6]) among the Mozambican strains. The P[8] strains grouped in two different lineages: P[8]-3 and P[8]-4, with totality of strains clustering together with P[8]-3 strains detected worldwide. P[8]-3 seems to be the only lineage currently circulating in different countries after the introduction of the massive anti-RVA vaccination (Rotarix® and/or RotaTeq®) worldwide Hemming and Vesikari [2013] and Silva et al. [2015] also reported the circulation of P[8]-3 in association with G1 in Finland and Brazil, respectively, for a period of over 20 years. Similar results were reported by Imbert-Marcille et al. [2013] showing a wide circulation of P[8]-3 associated with different G genotypes in 62 patients with diarrhea in France during 2010–2012. The MOZ21162 grouped inside the P[8]-4 lineage, a relatively recent lineage of P[8] reported in different parts of Europe, Africa, and Asia [Zade et al., 2009; Tamura et al., 2010; Zhirakovskaia et al., 2012; Zeller et al., 2015]. Since information on P[8]-4 sequence is rather limited in current available literature the effectiveness of the two RVA vaccines available worldwide against this lineage is not known.
The MOZ21197 strain grouped in the P[4] genotype, clustering together with G2P[4] and G8P[4] human prototype strains detected worldwide. Since its G genotype could not be characterized, it is not possible to affirm its DS-1 genogroup pattern. Further analysis will be required to obtain this information.
The three P[6] Mozambican strains grouped in the P[6]-I lineage. The MOZ2178 (G2P[6]) and MOZ21168 (G12P[6]) clustered together with human prototype strains detected in Bangladesh, Burkina Faso, Germany, India, South Africa, and USA between the years 2000 and 2010. The MOZ21205 strain (G4P[6]) grouped in a different cluster of P[6]-1, with porcine and human porcine-like strains, corroborating the VP7 gene result, which showed close relation with G4 human porcine-like strains detected in China, India, and Thailand. These results suggest the resorting character of this Mozambican strain, detected from a diarrheic child of 5 months living in Chokwe District. The rural population participating in this study was living close to animals such as goats, cattle, cats, dogs, ducks, and chickens, sharing all the same water source. Unfortunately, it was not possible to collect animalś specimens to compare with our data. Further analysis will be performed to confirm this hypothesis.
In the present study, the presence of the most frequently detected genotypes worldwide as G1, G3, G4, and G9, circulating in low percentages in Mozambique was identified. These findings are concordant with previous studies reporting the high detection of RVA genotypes co-circulating in Africa [Leite et al., 2008; Esona et al., 2009; Seheri et al., 2010; Waggie et al., 2010; Cunliffe et al., 2012; Doro et al., 2014; Seheri et al., 2014]. From our knowledge, this is the first data on molecular characterization of RVA in Mozambique showing the importance of RVA infection in children ≤2 years old as well as the diversity of RVA circulating in the Southern region of the country. Previous studies conducted in sub-Saharan Africa have demonstrated RVA diversity as one of the aspects related with the low efficacy of the vaccines [Madhi et al., 2010] although both vaccines are considered to confer heterotopic protection [Pitzer et al., 2011]. Therefore, the epidemiological data on RVA strains is important to characterize the detection frequency of different circulating RVA genotypes in order to identify possible changes in the epidemiological profile of the genotype dynamics that can influence the effectiveness of anti-RVA vaccination programs. Considering that the RV1 vaccine was introduced in the NIP of Mozambique in September 2015, the results obtained in the present study emphasizes the need to establish a national surveillance system to monitor the dynamics of RVA genotypes circulation in the country.
ACKNOWLEDGMENTS
We would like to thank all the children and their guardians WHO accepted to take part in this study. We extended our gratitude to all clinical staff at Chókwè Rural Hospital, Chókwè Health Centre, Carmelo Hospital and to the Laboratory of Comparative and Environmental Virology, Oswaldo Cruz Institute, Fiocruz, Brazil for their support during fieldwork and laboratory procedures.




