Porcine 2′, 5′-oligoadenylate synthetases inhibit Japanese encephalitis virus replication in vitro
Abstract
The 2′, 5′-oligoadenylate synthetases (OAS) are antiviral proteins and several isoforms have been identified as flavivirus-resistance biomarkers in human and mouse. The expression kinetics and antiviral functions of porcine OAS family (OAS1, OAS2, and OASL) in PK-15 cells following infection by Japanese encephalitis virus (JEV) were evaluated in the present study. The endogenous expression of the three OAS genes was efficiently induced by IFN-α treatment in PK-15 cells. However, expression of pOAS1 and pOAS2 responded more quickly than pOASL. Infection by JEV also induced the expression of the pOAS isoforms, but at a significantly lower level than that observed following IFN-α stimulation. Transient overexpression of pOASL and pOAS1 inhibited JEV replication more efficiently than OAS2 overexpression. Interestingly, knockdown of pOAS2 expression by siRNA treatment led to the highest increase in JEV multiplication. Co-silencing of RNase L and each pOAS revealed that the anti-JEV function of pOAS1 and pOAS2 were RNase L dependent, while the antiviral activity of pOASL was not. In conclusion, all pOAS isoforms play a significant role in the response to JEV infection, and are differentially induced by different stimuli. The alternative pathways of antiviral activity stimulated by OASL require further study. J. Med. Virol. 88:760–768, 2016. © 2015 Wiley Periodicals, Inc.
Abbreviations
-
- DENV
-
- dengue virus
-
- JEV
-
- Japanese encephalitis virus
-
- JE
-
- Japanese encephalitis
-
- m.o.i.
-
- multiplicity of infection
-
- mRNA
-
- messenger RNA
-
- OAS
-
- 2′, 5′-oligoadenylate synthetase
-
- OASL
-
- OAS-like
-
- rRNA
-
- ribosomal RNA
-
- si
-
- small interfering
-
- WNV
-
- West Nile virus
INTRODUCTION
Japanese encephalitis virus (JEV) is an arbovirus belonging to family Flaviviridae, genus Flavivirus. This virus is maintained in a zoonotic cycle involving pigs, ardeid birds, and Culex species of mosquitoes, with humans being dead-end hosts [Unni et al., 2011]. Japanese encephalitis (JE) caused by JEV is one of the most important viral encephalitides in Eastern and Southeastern Asia, affecting over 50,000 patients and resulting in 15,000 deaths annually [Misra and Kalita, 2010], Those affected by JE are primarily children and, therefore, JE is considered a major public health problem. It is well-known that JE also influences the health and breeding of pigs, which are the major amplifying host of JEV in nature [van den Hurk et al., 2009].
The 2′, 5′-oligoadenylate synthetases (OAS) belong to a highly conserved family that shares no significant overall sequence homology with other interferon-stimulated proteins. The OAS family consists of four genes encoding four proteins with different isoforms: OAS1 (isoforms p42, p44, p46, and p48), OAS2 (isoforms p69 and p71), OAS3 (isoform p100), and OASL (OAS-like protein; isoform p59). The OAS1, OAS2, and OAS3 enzymes contain one, two, and three repeats of the basal OAS functional unit, respectively. In contrast, OASL contains one OAS unit, a C-terminal domain homologous to a tandem ubiquitin repeat, and lacks 2′, 5′-oligoadenylate synthetase activity for characteristic changes in the active site [Hartmann et al., 1998; Justesen et al., 2000; Hartmann et al., 2003; Kristiansen et al., 2011]. The OAS2 and OAS3 proteins are only present in mammals, while OAS1 and OASL are widely distributed [Kjaer et al., 2009].
Porcine OASs show highly sequence homology to those expressed in human and mouse. The porcine OAS family also consists of OAS1, including two splice variants (OAS1a and OAS1b) that share 89.1% amino acid sequence identity, OAS2, and OASL, while OAS3 is absent [Perelygin et al., 2006]. The first OAS to be crystallized successfully was porcine OAS1, which is 73% identical in sequence to human OAS1 and has similar enzymatic characteristics [Hartmann et al., 2003].
As antiviral proteins, the OASs are induced by type I interferon and are an important component of the mammalian innate immune system [Hovanessian, 2007]. The OAS proteins are characterized by their ability to catalyze the synthesis of 2′, 5′-linked oligoadenylates with the general formula pppA(2′p5′A) n (n > 1) and are also sometimes referred to as the 2-5A synthetases [Roberts et al., 1976; Hovanessian et al., 1977; Kerr et al., 1977; Kerr and Brown, 1978; Hovanessian and Kerr, 1979]. The synthesis of intracellular 2′, 5′-linked oligoadenylates results in the activation of RNase L and the degradation of RNA [Kristiansen et al., 2011]. Activated RNase L degrades both viral and cellular RNAs, including rRNAs and mRNAs, with little sequence specificity (typically after UU or UA sites), which results in the inhibition of global protein synthesis and the suppression of viral replication during infection [Hovanessian, 2007; Kwon et al., 2013].
Numerous studies have demonstrated the antiviral effects of OAS proteins. The flavivirus resistance (Flvr) gene was identified as OAS1b simultaneously by two groups that observed a restriction of West Nile virus (WNV) replication in wild mice by OAS1b [Mashimo et al., 2002; Perelygin et al., 2002]. It was subsequently determined that the flavivirus resistance conferred by mouse OAS1b was not mediated by the 2-5A/RNase L pathway [Elbahesh et al., 2011; Courtney et al., 2012]. Lin et al. [2009] demonstrated that human OAS1 (p42/p46) and OAS3 (p100) possessed antiviral activity against dengue virus (DENV). Kwon et al. [2013] verified that OAS1 (p46) and OAS3 (p100) defense against the hepatitis C virus (HCV) was dependent upon RNase L. A single nucleotide polymorphism (SNP) in the OAS1 gene was also associated with an increased risk of WNV disease in humans [Lim et al., 2009]. Furthermore, extracellular OAS1 entered into cells and possessed strong antiviral activity in both in vitro and in vivo studies; however, this activity was independent of RNase L [Kristiansen et al., 2010] and was found to have its affect at an earlier step in the viral replication cycle [Thavachelvam et al., 2015].
Japanese encephalitis virus and WNV are very closely related and are included, along with DENV, in the genus Flavivirus. Although the anti-WNV activities of human and murine OAS proteins have been described, the effect of porcine OAS proteins on JEV infection remains unclear. In the present study, the antiviral characteristics of porcine OAS members against JEV were systematically evaluated in vitro. The results indicate that porcine OASs have non-redundant antiviral activity and respond to different kinds of stimulation.
MATERIALS AND METHODS
Cell Lines and Viruses
Porcine kidney cells (PK-15) were grown in RPMI 1,640 medium (GIBCO™, Invitrogen, USA) supplemented with 10% heat inactivated fetal bovine serum (FBS). Baby hamster kidney cells (BHK-21) for the plaque forming assay were grown in Dulbecco's modified Eagle's medium (GIBCO™, Invitrogen) containing 10% FBS. The cultures were incubated at 37°C under 5% CO2. Japanese encephalitis virus strain NJ 2008 (GQ918133.2) was used for viral infection.
Plasmids and Porcine Interferon Treatment
The OAS genes (OAS1a: NM214303.1; OAS1b: AY550259.1; OAS2: AY288913.1; OASL: NM001031790.1) were cloned from PK-15 cells. The p3xFLAG-CMV™-7.1 expression vector (Sigma–Aldrich, USA) was used to construct pFLAG-OAS1a, pFLAG-OAS1b, pFLAG-OAS2 and pFLAG-OASL.
Porcine interferon alpha (IFN-α) was expressed in Pichia pastoris X-33 and purified by filtration and dialysis as previously described [Cao et al., 2009, 2006]. The PK-15 cells were treated with 1000 IU/ml porcine IFN-α diluted in cellular growth medium for indicated time. After treatment, the cells were harvested and subjected to qRT-PCR to detect OAS and GAPDH mRNA using specific primers (Table I).
| Primer | Nucleotide sequence (5′→3′) |
|---|---|
| JEV | Forward-CCTCCGTCACCATGCCAGTCTTAG |
| Reverse-TTCGCCATGGTCTTTTTCCTCTC | |
| GAPDH | Forward-TGGTGA AGGTCG GAGTGA AC |
| Reverse-GGA AGATGGTGATGGGATTTC | |
| OAS1 | Forward-AGAGTCCACGACGGGAGAACC |
| Reverse-ACTGACCCAGGGCATCAAAGG | |
| OAS2 | Forward-TCCGCCATTCGGCTACAAAG |
| Reverse-CCTGGGAGCCTTCCATTTTG | |
| OASL | Forward-CCTATGGCTACAGATGGGAC |
| Reverse-GGACTGGGCTCTTGTTGTT | |
| RNase L | Forward-GATTGCTGGTCCTCTATGTG |
| Reverse-AGGGTCCTGTAACGGCTC | |
| IFN-alpha | Forward-TGGTGCATGAGATGCTCCA |
| Reverse-GCCGAGCCCTCTGTGCT | |
| OAS1 siRNA | GAGAAUUCAUCCAGGAAAU |
| AUUUCCUGGAUGAAUUCUC | |
| OAS2 siRNA | CCACCAAACUGAAGGAUUU |
| AAAUCCUUCAGUUUGGUGG | |
| OASL siRNA | AGGUGGAGCUGGUGGCAUU |
| AAUGCCACCAGCUCCACCU | |
| RNase L siRNA | UGGAAGAGAUGAAUGCAUA |
| UAUGCAUUCAUCUCUUCCA |
Cell Transfection and Viral Infection
The PK-15 cells were seeded into 24-well plates with antibiotic-free medium. When the cell monolayer reached 80–90% confluence, 500 ng of each eukaryotic expression vector (pFLAG-OAS1a, pFLAG-OAS1b, pFLAG-OASL, and pFLAG-OAS2) or negative control vector (p3xFLAG-CMV™-7.1) were individually transfected into cells in Opti-MEM™ medium (Invitrogen) containing Lipofectamine®3000 Reagent (Invitrogen) according to the manufacturer's instructions.
The PK-15 cells were transfected with 40 nM of OAS1 siRNA, OAS2 siRNA, OASL siRNA, RNaseL siRNA or scrambled (nonsense) siRNA (designed and synthesized by Shanghai Novland, China) for 48 hr. After transfection, cells were infected with JEV (0.1 m.o.i.) for another 24 hr, followed by detection of JEV-specific RNA and OAS or RNase L mRNA by qRT-PCR. For the co-transfection experiment, 20 nM OAS siRNA and 20 nM RNase L siRNA were used as described above.
Real-Time Quantitative Polymerase Chain Reaction (PCR)
Total RNA was extracted from the PK-15 cells using TRIzol reagent (TaKaRa, Dalian, China). Reverse transcription was performed using PrimeScript™ RT Master Mix (TaKaRa) following the manufacturer's protocol. Briefly, 500 ng of total RNA, 2 µl of 5xPrimeScript™ RT Master Mix, and RNase free water were combined in a total volume of 10 µl in a nuclease-free microcentrifuge tube. The mixtures were heated to 37°C for 15 min and quickly inactivated at 85°C for 5 sec to remove RNA complementary to cDNA. Real-time quantitative PCR was performed using the SYBR quantitative real-time PCR system (TaKaRa) following the manufacturer's protocol. The specific primers used are listed in Table I. Viral PCR results are presented as the numbers of RNA viruses. The expression of IFN-α and OAS mRNA was normalized to the expression of GAPDH as the endogenous control. The relative changes in gene expression were calculated using the 2−ΔΔCT method [Schmittgen and Livak, 2008].
Immunoblotting Assay
Cells were harvested, washed with PBS three times, and lysed in RIPA buffer (10 mM Tris–HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 0.1% SDS, 0.25% sodium deoxycholate, and 1 mM PMSF). The samples were analyzed by 12% SDS-PAGE and transferred onto nitrocellulose membranes (Bio-Rad, USA). After washed and blocked, the blots were incubated with primary antibodies, including monoclonal anti-NS1′ antibody, monoclonal anti-Flag (Genescript, Nanjing, China) or anti-β-actin antibody (Sigma–Aldrich), diluted in TBST buffer (10 mM Tris–HCl [pH 8.0], 150 mM NaCl, 0.5% Tween 20) containing 5% skim milk at 4 °C overnight. The membranes were incubated with secondary antibody (horseradish peroxidase conjugated goat anti-mouse IgG; Santa Cruz, USA) diluted in TBST buffer containing 5% skim milk for 1 hr at room temperature. And the membranes were washed thrice with TBST and the bands were detected using ECL reagents (Tanon, Shanghai, China).
Plaque Forming Assay
Serial dilutions of the supernatants from transfected and infected PK-15 cells were used to inoculate BHK-21 cells grown in 6-well plates for 1.5 hr. Overlay medium (2% low melting-point agarose with DMEM medium containing 2% FBS) was added to each well, and the plates were incubated for another 72 hr at 37°C under 5% CO2. The cells were then stained with 0.5% crystal violet and plaques were counted.
Immunofluorescence Assay
The PK-15 cells were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS), permeabilized with 0.5% Triton X-100, and incubated for 1 hr in blocking buffer (PBS containing 5% BSA). The cells were incubated at 4°C overnight with mouse monoclonal anti-NS1′ antibody in blocking buffer (1:1,000), washed with PBS, and then incubated for 2 hr at room temperature in blocking buffer containing rabbit anti-FLAG polyclonal antibody (1:500; Genescript), washed with PBS, and then incubated with fluorescein-(FITC) labeled goat anti-mouse IgG (1:100; KPL) and rhodamine-labeled goat anti-rabbit IgG (1:100; KPL). The cells were stained with 1 µg/ml DAPI (Boster, Wuhan, China) in PBS for 10 min and washed with PBS. Images were obtained using a Carl ZEISS inverted fluorescence microscope.
Statistical Analyses
All of the assays described were repeated at least three times and all measurements were performed in triplicate. Mean and standard deviation (mean ± SD) values were calculated using Microsoft Excel. The figures and statistical analyses were performed using GraphPad™ Prism 5.0 software. Statistical significance (*P < 0.05, **P < 0.01) and non-significant differences were determined using a two-tailed Student's t-test.
RESULTS
OAS mRNA Expression in PK-15 Cells Is Induced by Porcine IFN-α and JEV
After 6 hr of incubation with 1,000 IU/ml porcine IFN-α, OAS1 and OAS2 mRNA expression rapidly increased over 62.2-fold, whereas the OASL expression was only increased 6.1-fold (Fig. 1A). The peak of mRNA expression for each of the OAS genes occurred at 24 hr post-infection. Increases of 119.1-, 84.6-, and 88.4-fold over controls were observed for OAS1, OAS2, and OASL, respectively. Slight decreases in OAS1 and OAS2 expression, and a larger decrease in OASL expression, were observed at 48 hr post IFN-α treatment.
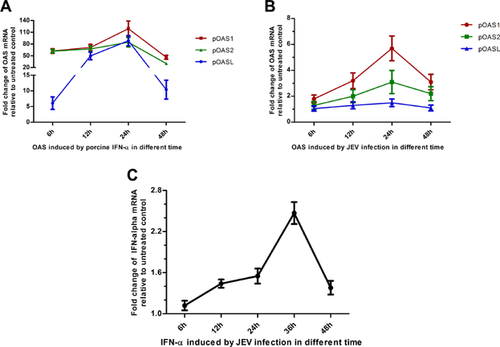
The expression kinetics of OAS isoforms in PK-15 cells stimulated by JEV were also investigated (Fig. 1B). The highest level of OAS expression following JEV treatment was observed after 24 hr. The stimulation of OASL expression by JEV infection was minimal, resulting in only a 1.5-fold increase. However, 5.7- and 3.1-fold increases were observed in JEV-induced OAS1 and OAS2 expression, respectively. Furthermore, OAS1 expression was always higher than that of OAS2 and OASL.
Only a low-level induction of IFN-α mRNA expression was observed following JEV infection (Fig. 1C).
OAS1a and OASL Over Expression Inhibits JEV Replication in PK-15 Cells
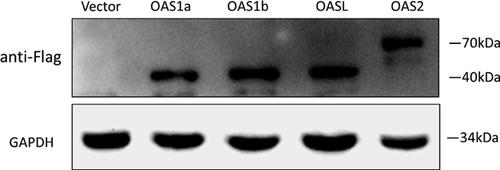
Immunoblotting revealed that OAS1a, OAS1b, OASL, and OAS2 were successfully expressed in PK-15 cells transfected with the pFLAG-OAS1a, pFLAG-OAS1b, pFLAG-OAS2, and pFLAG-OASL expression vectors, respectively (Fig. 2). When compared to the detected levels of GAPDH, a similar efficiency of transfection was observed among the OAS expression constructs.
To assess the anti-JEV activity of each porcine OAS isoform, PK-15 cells were transfected with the pOAS constructs, infected with JEV 24 h later, and JEV genome abundance was quantified 24 h post inoculation. In contrast to the vector control, the relative ratio of JEV genome was 0.14 (P < 0.01), 0.75 (P > 0.05), 0.47(P < 0.05) and 0.028 (P < 0.01) in PK-15 cells transfected with the pOAS1a, pOAS1b, pOAS2, and pOASL constructs, respectively (Fig. 3A). The decreases in JEV genome abundance in the pOAS1 and pOASL groups were also statistically significant compared to that in the pOAS2 group (P < 0.01). In addition, the supernatants were collected from the treated PK-15 cells at 24 hr post inoculation to compare JEV titers using a plaque forming assay at a dilution of 1:1000 (Fig. 3B). The plaque forming assay demonstrated that the viral titers in the supernatants from the pOAS1a-, pOAS2-, and pOASL-transfected groups were significantly reduced relative to the control, but that the level in the pOAS1b-transfected group was similar to the vector control (Fig. 3C).
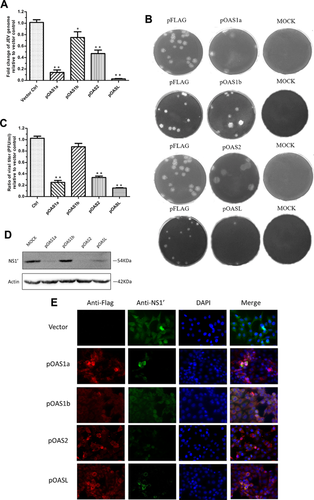
A monoclonal anti-JEV NS1′ antibody developed in this laboratory was used for immunoblotting the JEV-treated PK-15 cells. The anti-NS1′ mAb is specific against the region of NS1′ ribosomal frameshifting and lacks cross-reaction with the NS1 protein [Meng et al., 2013]. No significant NS1′ protein band was observed in the pOAS2-transfected group, and only faint bands were observed in the pOAS1a- and pOASL-transfected groups. Distinct bands were observed in the pOAS1b-transfected and vector control groups (Fig. 3D).
Furthermore, the antiviral effects of porcine OAS were evaluated using an indirect immunofluorescence assay to detect viral antigen and OAS expression in treated cells (Fig. 3E). To obtain clearer images, the transfected PK-15 cells were infected with JEV at 1 m.o.i.. In cells transfected with OAS1a, OAS2, and OASL, NS1′ protein specific florescence was significantly reduced, implying a strong inhibition of JEV replication, while the effect of OAS1b was not significant.
OAS2 Knockdown Leads to Increased JEV Replication in PK-15 Cells
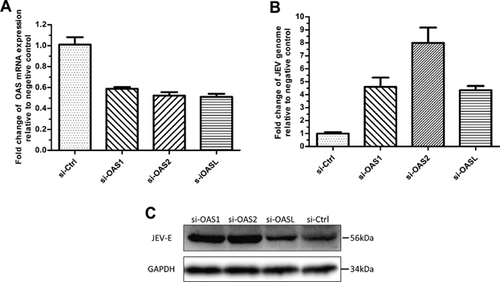
Knockdown of the OAS isoform in PK-15 cells by siRNA was used to assess whether each porcine OAS isoform is necessary for the defense against JEV. The results of qRT-PCR and immunoblotting (Fig. 4B and C, respectively) indicate that the quantity of JEV genomes increased significantly in OAS knockdown cells. Notably, silencing of the OAS2 gene resulted in up to a 7.99-fold increase in JEV genome abundance compared with the scrambled siRNA treatment. Knockdown of OAS1 and OASL resulted in approximately a 4.5-fold increase in JEV genome abundance. It is important to note that the efficiency of OAS gene knockdown for the three porcine OAS genes in PK-15 cells was not high (approximately 48%) and was not significantly different from the each other (Fig. 4A).
RNase L-Dependent (OAS1 and OAS2) and -Independent (OASL) Antiviral Activity
In PK-15 cells, a 32% knock down of RNase L mRNA by 40 nM RNase L siRNA facilitated a 5.22-fold increases in JEV replication (Fig. 5A and 5B). This increase in JEV replication was also evident in immunoblots for testing JEV envelope protein (Fig. 5C).
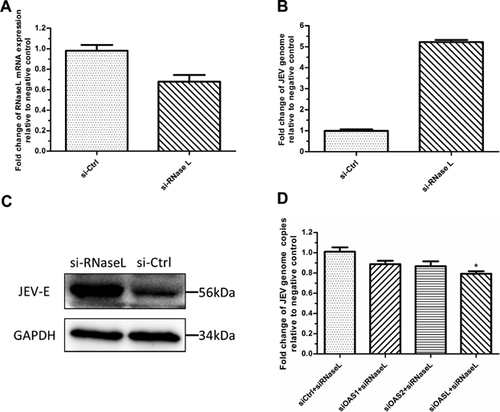
To assess whether the anti-JEV activity of each OAS protein depended upon the RNase L pathway, PK-15 cells were co-transfected with 20 nM OAS siRNA and 20 nM RNase L siRNA, followed by JEV infection (0.1 m.o.i.). The reduction in JEV genome abundance in the OASL/RNase L co-silence groups (79.12%) was statistically significant (P < 0.05) compared to the control group. However the reduction in the OAS1/RNase L (88.62%) and OAS2/RNase L (86.6%) co-silence groups were not significant (P > 0.05) (Fig. 5D).
DISCUSSION
In this study, the endogenous expression of OAS was efficiently stimulated by porcine IFN-α and JEV infection in a time-dependent manner. The replication of JEV in PK-15 cells was significantly reduced by overexpression of OAS1a and OASL proteins, and was enhanced by the knockdown of porcine OAS2 gene expression. These results suggest an antiviral role for each of the OAS isoforms during JEV infection. A major antiviral effect of porcine OAS1 is suggested during the early stage of JEV infection, as evidence by the rapid and efficient expression of this OAS induced by JEV and IFN-α. In addition, the high basal anti-JEV potential of OAS2 suggested that it is also important in the antiviral response. Furthermore, it was also observed that OASL depend upon alternative anti-JEV pathways, while OAS1 and OAS2 depend upon the classical OAS/RNase L pathway.
In PK-15 cells, OAS1 gene expression was stimulated by porcine IFN-α and JEV more efficiently and more rapidly than other OAS isoforms, especially within 24 hr post treatment. Furthermore, JEV replication was remarkably inhibited by the overexpression of porcine OAS1a at the mRNA and protein level. These results indicate that OAS1 might play a major role in the early stage of JEV infection, with the potential for high and rapid endogenous expression. The reasons for the lack of a rapid upregulation of OASL in PK-15 cells may relate to differences in the regulation of the expression of this protein, or to the different antiviral mechanisms stimulated by this OAS, as was observed in this study. These differences in expression following infection are the opposite of what has been observed for human OASL and OAS1. In addition, the lack of a significant increase in the expression of IFN-α following JEV infection in PK-15 cells may explain the relatively low level of induction of the expression of all OAS isoforms following JEV treatment.
Knockdown of OAS2 gene expression in PK-15 cells facilitated a significantly higher level of JEV replication compared to the knockdown of OAS1 and OASL gene expression. However, OAS2 overexpression was not as efficient as the overexpression of OAS1a and OASL in the suppression of viral replication. This may suggest that OAS2 possesses a higher level of basal anti-JEV potential. These results also reveal that the antiviral effects of OAS1a may be dose-dependent, as this OAS exhibited a more prominent anti-JEV activity following overexpression than OAS2, but a lower activity than OAS2 following knockdown. Furthermore, extracellular OAS1 has been demonstrated to have strong dose-dependent antiviral activities, although these activities were independent of RNase L [Kristiansen et al., 2010; Thavachelvam et al., 2015].
The antiviral activity of porcine OAS1b was not obvious in our study despite the fact that OAS1b is 89.1% identical in amino acid sequence to OAS1a, which demonstrated strong antiviral activity. It may be speculated that the difference between OAS1a and OAS1b in the observed anti-JEV effect is relative to the splice variants of OAS1, as the OAS1 gene contains two 3′-exons [Kristiansen et al., 2011]. Future studies should investigate why these two OAS1 isoforms possess different antiviral activities, as they both contain a basal OAS domain and only differed in their C termini.
An abundance of studies have demonstrated that OAS1 and OASL exhibit antiviral effects against various viruses through different pathways and mechanisms [Kwon et al., 2013; Lee et al., 2013a,2013b; Deo et al., 2014; Imran et al., 2014; Zhu et al., 2014]. Based on the results of the co-silencing of the OAS and RNase L genes, it was confirmed that porcine OAS1 and OAS2 function through the IFN-α/OAS antiviral pathway, and that OASL functions through some other antiviral pathway. A distinct effect of OASL has previously been related to its specific structure. Very recently, Zhu et al. [2014] described the antiviral mechanisms of human OASL, whereby OASL binds to and enhances retinoic acid inducible gene I (RIG-I) -mediated antiviral signaling through its C-terminal ubiquitin-like domain. In the current study, the porcine OASL exhibited robust anti-JEV activity and was nearly as effective as porcine OAS1. However, the porcine OASL mRNA sequence contains premature stop codons and encodes truncated proteins that lack the typical C-terminal double ubiquitin domains that are characteristic of human OASL. Further study of the antiviral mechanisms of the truncated porcine OASL proteins is required.
The present study has focused on the interaction between JEV and the porcine OAS proteins with the goal of confirming the antiviral activities of these proteins and providing information that could be used to prevent JEV infection in pigs. The antiviral activities of porcine OAS1, OAS2, and OSAL against JEV were demonstrated in vitro in PK-15 cells. Further study of these porcine OAS proteins is necessary to understand their antiviral mechanisms at the cellular level and to determine their antiviral effects in natural hosts of the virus.




