The interaction of a single-nucleotide polymorphism with age on response to interferon-α and ribavirin therapy in female patients with hepatitis C infection
Abstract
Older female patients exhibit a poor response to the current standard treatment for hepatitis C, interferon-α, and ribavirin (PEG-IFN-α/RBV). In this study, we reported that the combination of age and the genotype of a novel SNP can predict response to standard treatment (P = 7.31 × 10−8). The model incorporating genotype of the novel SNP, rs1287948, predicts response more accurately (AUC = 0.934; 95% CI = 0.881–0.988) in women as compared with the model using age and the previously identified SNP, rs8099917. J. Med. Virol. 86:1130–1133, 2014. © 2014 Wiley Periodicals, Inc.
INTRODUCTION
In late 2009, genome-wide association studies (GWAS) were used to identify two single-nucleotide polymorphisms (SNPs) located near the IL28B (IFNλ3) gene, rs12979860, and rs8099917, strongly associated with the response to pegylated-interferon-α plus ribavirin (PEG-IFN-α/RBV) therapy for hepatitis C; the identity of the SNPs and their association with treatment response has been independently verified by three separate groups [Ge et al., 2009; Suppiah et al., 2009; Tanaka et al., 2009]. The SNP genotypes are recognized as important pretreatment predictors of the response to PEG-IFN-α/RBV therapy, as described in the present guidelines for the management of hepatitis C [Ghany et al., 2011; Editors of the Drafting Committee for Hepatitis Management Guidelines: The Japan Society of Hepatology, 2013]. Since the identification of these SNPs, the molecular mechanism by which the IL28B (IFNλ3) gene influences the difference in response to PEG-IFN-α/RBV treatment has been investigated but has not yet been clarified. Recently a new dinucleotide variant near the IL28B gene ss469415590 (TT or ΔG), which is strongly linked to rs12979860 and rs8099917, has been discovered [Prokunina-Olsson et al., 2013]. Interestingly, the ΔG allele creates a novel interferon gene, IFNL4, and is thought to reduce responses to type I (including IFN- α) and type III interferons [Prokunina-Olsson et al., 2013]. Meanwhile, it is known that the response to PEG-IFN-α/RBV in older women is poor [Sezaki et al., 2009]. A genetic signature associated with the poor responder phenotype might exist and may be correlated with age. In this paper, we report that the result of a genome-wide interaction analysis between SNP status and age in female patients treated with PEG-IFN-α/RBV therapy. These results will lead to further improvement in prediction accuracy when assessing the probable response to Hepatitis C treatment and may also lead to further understanding of the molecular mechanism by which treatment response in altered older women.
MATERIALS AND METHODS
Study Patients
In a previous study conducted by our group, genotypes of 142 Japanese patients (67 male and 75 female) treated with PEG-IFN-α/RBV therapy were used in a GWAS [Tanaka et al., 2009]. The genotypes were derived from an Affymetrix SNP 6.0 array for 900,000 SNPs. The present study uses the same SNP data and the corresponding demographic information from the 75 women who were members of the original group of 142 patients. Of the 75 female patients, 29 patients exhibited a virologic response (VR) while 46 patients exhibited a null virologic response (NVR). A total of 672,883 SNPs on autosomal or X chromosomes met our criteria: SNP call rate ≥95%, minor allele frequency ≥1%, and a Hardy–Weinberg equilibrium P-value ≥0.001 for all 75 female patients; the identified SNPs were used for the subsequent analyses. As stated previously [Tanaka et al., 2009], informed consent was obtained from all patients and the study protocol conforms to the relevant ethical guidelines as reflected in a priori approval by the ethics committees of all the participating universities and hospitals.
Genome-Wide Interaction Analysis
In order to determine whether there was an effect modifier of relation between age and the response to PEG-IFN-α/RBV, a genome-wide interaction analysis (i.e., 672,883 SNP—age interaction tests) was performed using a logistic regression model. The genotype of each SNP was regarded as the number of alleles in each individual (i.e., 0, 1, or 2). The likelihood ratio test, which uses the deviance reduction due to including the SNP—age interaction as the χ2 test statistics, was employed.
Detailed Analyses for Detected Interaction
After the genome-wide interaction analysis, detailed analyses including a newly detected SNP, rs12879468, and/or age (see Model in Table I) were conducted using the logistic regression with the χ2—based likelihood test as described above.
| Model Logit (response) | Deviance | Model compared | Likelihood ratio | P-value | |
|---|---|---|---|---|---|
| Const. | 100.085 | ||||
| 1: | Geno | 99.719 | vs. Const. | 0.366 | 0.545 |
| 2: | Age | 92.854 | vs. Const. | 7.231 | 7.17 × 10−3 |
| 3: | Geno + age | 92.070 | vs. Model 2 | 0.784 | 0.376 |
| 4: | Geno + age + age × geno | 63.088 | vs. Model 3 | 28.982 | 7.31 × 10−8 |
- Geno, number of G allele for rs1287948 in individuals (0:GG,1:GC). Note that genotype CC does not exist in the sample.
To assess the improvement of the predictive performance by adding the rs1287948 SNP—age interaction to the model having age and rs1287948 as main effects (see Model 3 and 4 in Table I), receiver operator characteristic (ROC) curves based on the predicted probabilities and corresponding area under the curves (AUC) were used. The confidence intervals for each AUC and the P-value for the difference between the two ACUs were obtained using DeLong's method [DeLong et al., 1988].
It was also investigated whether prediction accuracy could be improved by adding the novel rs1287948 SNP to the model, which includes only age and the preexisting SNP, rs8099917 that was identified in previous studies [Suppiah et al., 2009; Tanaka et al., 2009]. That is, the two models: Logit = ext.Geno + Age (the existing model) versus Logit = ext.Geno + Age + Geno + Age × Geno, where Geno is the number of G alleles for rs1287948 and ext.Geno is the number of T alleles for rs8099917 in individuals, were compared with each other. For the analysis, ROC, and AUC were used and the confidence interval of each AUC and the P-value for the difference between two AUCs were obtained using DeLong's method [DeLong et al., 1988].
RESULTS
Using genome-wide interaction analysis, we was found that the rs1287948 SNP located on chromosome 1 exhibited a strong interaction with age (P = 7.31 × 10−8) regarding the response to PEG-IFN-α/RBV and reached genome-wide significance with a Bonferroni-corrected significance level of P = 7.43 × 10−8 (=0.05/672,883). No other SNP showed significant evidence of interaction. Furthermore, there was no interaction of rs1287948 SNP with age in 67 male patients (P = 0.48).
Table I presents the results of fitting a series of logistic regression models involving genotypes of rs1287948 and/or age. Note that rs1287948 could be found only when the interaction term (Age × Geno) was considered (see the P-values in the rows of Model 1, 3, and 4). Figure 1 shows the probability of the predicted response and the observed response to PEG-IFN-α/RBV therapy according to rs1287948 genotype. A clear SNP—age interaction for rs1287948 (i.e., the reverse effect of age in the two different genotypes, CG and GG, of rs1287948) can be seen.
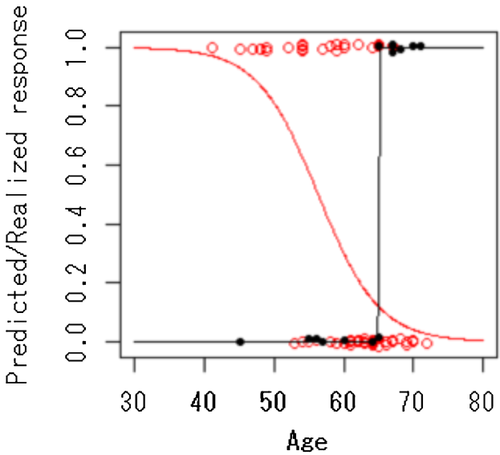
Figure 2 shows the AUCs for Model 3 and Model 4 in Table I. For the no-interaction model (Model 3), the AUC is 0.704 (95% confidence interval, CI = 0.586–0.822). Adding the interaction term (Model 4), the AUC was improved to 0.867 (95% CI, 0.784–0.949). The P-value for the difference between the two AUCs was P = 0.000826. Thus, the rs1287948 SNP—age interaction is considered to be an effective predictor of response to PEG-IFN-α/RBV therapy.
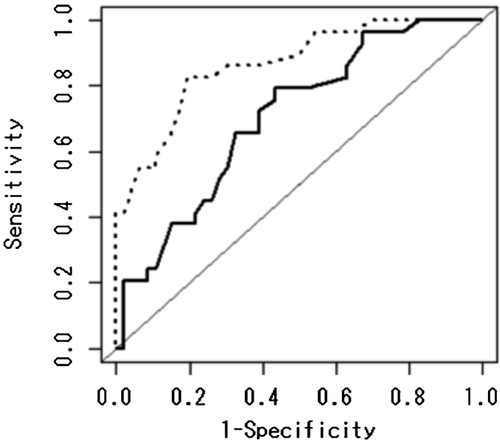
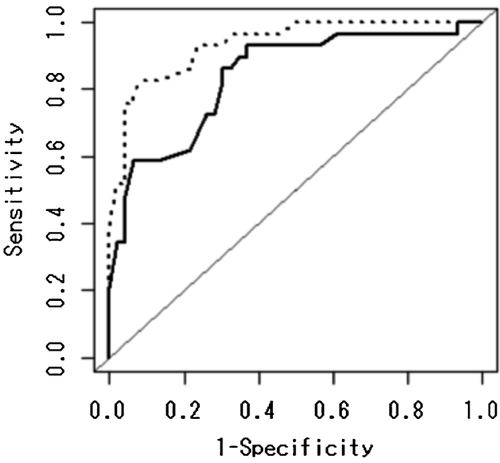
Figure 3 shows the improvement of the predictive performance by adding the rs1287948 SNP into the existing model including only age and the preexisting SNP, rs8099917 (for more details, see Material and Methods section). For the existing model, the AUC was 0.839 (95% CI, 0.744–0.933). After including the novel rs1287948 SNP, the AUC improved to 0.934 (95% CI, 0.881–0.988). The P-value for the difference between the two AUCs is P = 0.0167. Therefore, it was concluded that adding the rs8099917 and its interaction term with age improve the prediction accuracy compared with the existing model.
The rs1287948 SNP is located in a non-coding region of chromosome 1 and is 108 kbp downstream from the PARP1 gene. PARP1 is important for repairing single-strand breaks and, as such, PARP1 inhibitors are currently being investigated as potential anti-cancer agents in patients carrying BRCA mutations [Rouleau et al., 2010]. Therefore, the rs1287948 SNP—age interaction involvement in the response to PEG-IFN-α/RBV therapy could be related to the function of PARP1.
DISCUSSION
In this paper, the impact of the rs1287948 SNP—age interaction on the response to PEG-IFN-α/RBV therapy was investigated. Although the importance of the analysis of gene–gene (SNP-SNP) interaction is recognized, there are few practical or successful GWASs [Cordell, 2009; Wu et al., 2009; Bansal et al., 2010]. Statistically detecting the interaction between SNPs is difficult because there are a large number of combinations of SNPs. Therefore it is more practical to first identify those SNPs that show a strong marginal effect before testing the interactions between them. Using such methods, SNPs with interactive effects but without marginal effects are missed. Conversely, studies of the interactions between SNPs and demographic or clinical characteristics are easy to conduct; the exhaustive test of all characteristic values and SNPs interactions can be done without first identifying an SNP with a strong marginal effect; this is a merit of the analysis. In fact, the rs1287948 SNP was identified using tests for the SNP—age interaction and could not be found by its marginal effect (see Table I). In addition, GWAS is becoming an useful tool in pharmacogenomics and personalized medicine [Madian et al., 2012]. In GWAS for pharmacogenomics, testing interactions between SNPs and demographic or clinical characteristics can and should be done, as the information gained leads to more effective personalized medicine.
ACKNOWLEDGMENTS
The authors thank Yasuhito Tanaka and Takayuki Yamada for comments on the early stages of this work.




