High prevalence of HHV8 infection and specific killer cell immunoglobulin-like receptors allotypes in Sardinian patients with type 2 diabetes mellitus
Abstract
The development of type 2 diabetes is thought to involve both environmental, possibly infectious, and genetic factors. Recently, a high prevalence of human herpesvirus 8 (HHV8) infection was observed in type 2 diabetes patients, and specific killer cell immunoglobulin-like receptors (KIR) allotypes were associated to both increased susceptibility to herpesvirus infection and risk to develop diabetes. However, no clear gene-disease or virus-disease associations have been established. To investigate the possible interplay between HHV8 infection, KIR allotype and type 2 diabetes, virus prevalence and KIR genotype were analyzed by PCR in 168 patients affected by type 2 diabetes and 108 control individuals belonging to the Sardinian population. Results showed a significant increase of HHV8 prevalence in type 2 diabetes patients versus controls (57% vs. 17%, P < 0.001), and a significant increase of KIR2DL2/DS2 homozygosity in diabetes patients infected with HHV8 compared to uninfected ones (64% vs. 14%, P < 0.0001), resulting in a significant OR of 11.31. In addition, the analysis of the frequency of the KIR2DL2/DS2 receptor and its HLA-C1 ligand, accordingly to the status of HHV8 infection, showed a significant increased correlation between KIR2DL2/DS2, type 2 diabetes and HLA-C1C1 genotype in the type 2 diabetes patients infected with HHV8 compared to uninfected ones (62% vs. 15%, P < 0.0001, OR = 8.64). These findings provide preliminary evidence that HHV8 infection might be a cofactor for type 2 diabetes in a specific subset of genetically susceptible individuals, and suggest the possibility that such patients might have an impaired immune-mediated component contributing to the development of type 2 diabetes. J. Med. Virol. 86: 1745–1751, 2014. © 2013 Wiley Periodicals, Inc.
INTRODUCTION
Type 2 diabetes is a major cause of morbidity and mortality worldwide [Green et al., 2003; Stovring et al., 2003]. The susceptibility to type 2 diabetes is thought to be related to both genetic and environmental factors, possibly including infection by chronic stressors for immune system such as herpesviruses. In accord with this hypothesis, we and others observed an increased frequency of human herpesvirus 8 (HHV8) infection in type 2 diabetes patients, respectively in the Sardinian population [Ingianni et al., 2007] and in the sub-Saharan Africans [Sobngwi et al., 2008].
HHV8, also known as Kaposi's sarcoma associated herpesvirus (KSHV), belongs to the Herpesviridae family, subfamily Gamma-herpesvirinae, genus Rhadinovirus. HHV8 is etiologically associated to all the clinical forms of Kaposi's sarcoma (KS), and is correlated to other disorders developing in immunosuppressed subjects [Schulz, 2000; Mesri et al., 2010], where it results highly tumorigenic [Wen and Damania, 2010]. Its prevalence varies geographically, and some studies have reported an increasing presence of HHV8 infection in the general population and in particular in some countries, such as Southern Italy, Sicily, Sardinia, and the Po river valley. Genetic pressure, caused by endemic diseases, has been claimed by some authors as being the possible cause of the selection of a population with an increased sensitivity to HHV8 infection [Coluzzi et al., 2002].
However, only a small fraction of individuals infected with HHV8 develop HHV8-associated tumors and/or other clinically relevant diseases, suggesting that other risk factors, likely acting as cofactors, are involved in HHV8-associated disease development. Interestingly, type 2 diabetes patients were observed to have an increased occurrence of KS [Gill and Shah, 2006]. Type 2 diabetes disease leads to a decrease in the number of T lymphocytes, depresses killer cell activity and decreases human resistance to herpesvirus infections [Guttman-Yassky et al., 2006]. A diminished resistance to herpesvirus infections was also observed in an experimental model of diabetes in mice [Kataoka et al., 1983].
On the other hand, the genetic background seems to play an equally important role in the risk to develop type 2 diabetes, and specific killer cell immunoglobulin-like receptors (KIR) present on NK cells have been reported to be associated with diabetes. In particular, the KIR2DL2 gene was suggested to be associated with the risk to develop diabetes [Ramos-Lopez et al., 2009], and was observed to be involved in the increased susceptibility to herpesvirus infection in multiple sclerosis, by altering NK cell response [Rizzo et al., 2012].
Based on these observations, the present study aimed at analyzing the possible interplay between KIR allotype, HHV8 infection and type 2 diabetes in patients of Sardinian descent.
MATERIALS AND METHODS
Study Patients
The study examined a group of 168 patients with a diagnosis of type 2 diabetes and 108 control subjects. All the subjects enrolled in the study, both type 2 diabetes patients and controls, belonged to the autochthonous Sardinian population. Type 2 diabetes group included 88 males and 80 females (mean age 65 ± 12 years, range 42–87 years). Control healthy subjects included 74 males and 34 females (mean age 44.4 ± 12 years, range 20–69 years). Exclusion criteria for enrollment in the study were: concomitance of KS or other HHV8-related diseases.
Type 2 diabetes patients were diagnosed at the Diabetes and Metabolic Diseases Service (Hospital S. Giovanni di Dio, AOU Cagliari), accordingly with criteria of the World Health Organization for classification of diabetes [WHO, 2006]. Blood samples were collected after receiving informed consent, and were used after approval of the Local Ethical Committee for analyses, to which purpose the samples were anonymized.
HHV8 DNA Detection
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood by Lymphoprep technique (Nycomed Pharma AS, Oslo, Norway). Total cellular DNA was extracted (Easy-DNA, Invitrogen, San Diego, CA) and analyzed by PCR amplification for HHV8 ORF26 gene, as already described [Ingianni et al., 2007]. The amplification products were detected by ethidium bromide–stained agarose gel electrophoresis. The PCR product specificity was confirmed by Southern blot assay, and the integrity and efficiency of the extracted DNA was checked by amplification of the human β-globulin gene [Ingianni et al., 2007].
KIR and HLA Class I Typing
Genotyping for 14 KIR genes was performed by KIR-typing kit (Olerup SSP KIR Genotyping; Olerup SSP AB, Saltsjobaden, Sweden), on total DNA extracted from PBMCs. To confirm the results obtained for KIR2DL2/DS2 and KIRD2DL3, DNAs were analyzed by PCR using specific primers and conditions, as previously described [Uhrberg et al., 1997]. Typing of HLA-C1 (HLA-C Asn80) and HLA-C2 (HLA-C Lys80) was performed by PCR, as described [Du et al., 2007].
The amplification products were analyzed by ethidium bromide–stained agarose gel electrophoresis.
Statistical Analysis
Statistical analysis was performed using the Stat View software package (SAS Institute, Inc., Cary, NC). Comparative analysis between study groups were performed by the Fisher exact text and χ2 test, and the Bonferroni correction for multiple comparison. P-values <0.005 were considered statistically significant (two-tailed).
RESULTS
HHV8 Presence in Type 2 Diabetes Patients
The results of HHV8 search (Fig. 1A) showed that 17% (19/108) of healthy donors were HHV8-positive, confirming previous data on HHV8 general prevalence in Sardinian population [Ingianni et al., 2007]. By contrast, 96/168 (57%) of type 2 diabetes patients resulted infected with HHV8, confirming the high prevalence of HHV8 infection in Sardinian patients affected by type 2 diabetes [Ingianni et al., 2007].
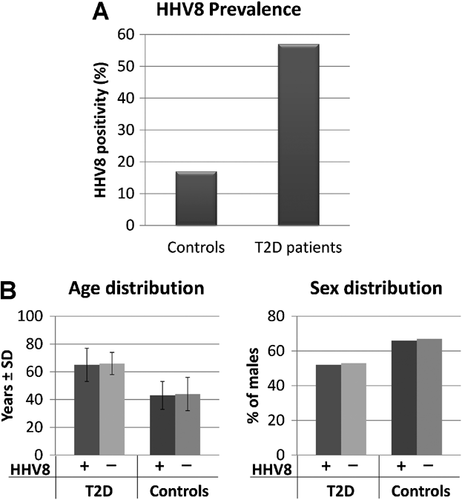
HHV8-positivity was unrelated to increasing age or sex prevalence, since HHV8-positive type 2 diabetes patients were 48% females and 52% males (mean age 65 ± 12 years), and HHV8-negative type 2 diabetes patients were 47% females and 53% males (mean age 66 ± 8 years) (Fig. 1B). Similarly, also HHV8-positive healthy donors were not significantly different from HHV8-negative individuals as to the mean age and sex distribution (respectively 43 ± 10 vs. 44 ± 12 years, and 66% vs. 67% males).
KIR Typing
Genotypization for 14 KIR genes, performed on the same PBMC DNA samples used for HHV8 detection, showed that the frequency of KIR genes frequency was similar in type 2 diabetes group versus controls, except for a decrease in KIR2DL3 (58% vs. 93%, Pc < 0.0001) (Table I). By contrast, when type 2 diabetes group was subdivided according to HHV8 infection, patients infected with HHV8 presented a significantly higher frequency of KIR2DL2/DS2 genes (considered together because of their strong linkage disequilibrium) compared with both uninfected type 2 diabetes patients (81% vs. 44%, Pc < 0.0001), and HHV8-infected (81% vs. 42%, Pc < 0.0001) or uninfected controls (81% vs. 53%, Pc < 0.0001). Conversely, KIR2DL3 gene frequency was lower in type 2 diabetes patients infected with HHV8 compared to HHV8-negative patients (36% vs. 86%, Pc < 0.0001) and to HHV8-infected controls (36% vs. 100%, Pc < 0.0001) or uninfected controls (36% vs. 78%, Pc < 0.0001).
| KIR genes | Total | T2D | CTR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T2D, N = 168 (%) | CTR, N = 108 (%) | P | Pc | HHV8(+), N = 96 (%) | HHV8(−), N = 72 (%) | P | Pc | HHV8(+), N = 19 (%) | HHV8(−), N = 89 (%) | P | Pc | |
| 2DL1 | 134 (80) | 85 (79) | 0.87 | NS | 78 (81) | 56 (78) | 0.69 | NS | 15 (79) | 70 (79) | 1 | NS |
| 2DL2 | 110 (65) | 55 (51) | 0.017 | NS | 78 (81) | 32 (44) | 1.3x10-6 | 1.8x10-5 | 8 (42) | 47 (53) | 0.45 | NS |
| 2DL3 | 97 (58) | 100 (93) | 5 × 10−11 | 7 × 10−10 | 35 (36) | 62 (86) | 4.2 × 10−11 | 5.8 × 10−10 | 19 (100) | 69 (78) | 0.35 | NS |
| 2DL4 | 168 (100) | 108 (100) | NS | NS | 96 (100) | 72 (100) | NS | NS | 19 (100) | 89 (100) | NS | NS |
| 2DL5 | 72 (43) | 45 (42) | 0.9 | NS | 39 (41) | 33 (46) | 0.53 | NS | 7 (37) | 38 (43) | 0.79 | NS |
| 2DS1 | 54 (32) | 32 (30) | 0.69 | NS | 31 (32) | 23 (32) | 1 | NS | 6 (32) | 26 (29) | 1 | NS |
| 2DS2 | 110 (65) | 55 (51) | 0.017 | NS | 78 (81) | 32 (44) | 1.3 × 10−6 | 1.8 × 10−5 | 8 (42) | 47 (53) | 0.45 | NS |
| 2DS3 | 34 (20) | 22 (20) | 1 | NS | 19 (20) | 15 (21) | 1 | NS | 4 (21) | 18 (20) | 1 | NS |
| 2DS4 | 118 (70) | 76 (70) | 1 | NS | 66 (69) | 52 (72) | 0.73 | NS | 14 (74) | 62 (70) | 1 | NS |
| 2DS5 | 50 (30) | 30 (28) | 0.79 | NS | 29 (30) | 21 (29) | 1 | NS | 5 (26) | 25 (28) | 1 | NS |
| 3DL1 | 168 (100) | 108 (100) | NS | NS | 96 (100) | 72 (100) | NS | NS | 19 (100) | 89 (100) | NS | NS |
| 3DL2 | 168 (100) | 108 (100) | NS | NS | 96 (100) | 72 (100) | NS | NS | 19 (100) | 89 (100) | NS | NS |
| 3DL3 | 168 (100) | 108 (100) | NS | NS | 96 (100) | 72 (100) | NS | NS | 19 (100) | 89 (100) | NS | NS |
| 3DS1 | 87 (52) | 57 (53) | 0.9 | NS | 49 (51) | 38 (53) | 0.88 | NS | 10 (53) | 47 (53) | 1 | NS |
- P, P-value obtained with Fisher exact test; Pc, P-value corrected for the number of comparisons.
Even more interesting, the analysis of KIR genotype (Table II) showed a strong difference between type 2 diabetes and controls, with an increase of KIR2DL2/DS2 homozygosity (42% vs. 7%, P < 0.0001), mainly due to type 2 diabetes patients infected with HHV8 in comparison with uninfected ones (64% vs. 14%, Pc < 0.0001), resulting in a significant OR of 11.31 (95% CI: 5.14–24.86). By contrast, none of the HHV8-positive controls was homozygous for KIR2DL2/DS2 genes, being either heterozygous (8/19) or homozygous for KIR2DL3 gene (11/19).
| KIR genotype | Total | T2D | CTR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T2D, N = 168 (%) | CTR, N = 108 (%) | χ2 | Pχ | HHV8(+), N = 96 (%) | HHV8(−), N = 72 (%) | χ2 | Pχ | HHV8(+), N = 19 (%) | HHV8(−), N = 89 (%) | χ2 | Pχ | |
| 2DL2-2DL2 | 71 (42) | 8 (7) | 62 (64) | 10 (14) | 0 (0) | 7 (8) | ||||||
| 2DL2-2DL3 | 39 (23) | 47 (44) | 40.1 | <0.0001 | 17 (17) | 22 (31) | 43.1 | <0.0001 | 8 (42) | 40 (45) | 1.88 | NS |
| 2DL3-2DL3 | 58 (35) | 53 (49) | 18 (18) | 40 (54) | 11 (58) | 42 (47) | ||||||
- χ2, Chi-squared test value; Pχ, P-value obtained with Chi-squared test.
Since the KIR2DL2/DS2 and KIR2DL3 receptors recognize HLA-C1 (HLA-C Asn80) as a specific ligand, the frequency of this gene was evaluated (Table III). Results showed a different distribution of HLA-C genotypes between type 2 diabetes patients and controls (χ2 = 5.59, P = 0.018) (48% vs. 31%, P = NS by Fisher test), which correlated to the increased HLA-C1C1 homozygosity in type 2 diabetes patients infected with HHV8 versus uninfected in the KIR2DL2/DS2-positive group (62% vs. 15%, P < 0.0001). Conversely, in KIR2DL3-group, HLA-C1C2 was more frequent in uninfected vs HHV8-infected type 2 diabetes patients (50% vs. 11%, P < 0.0001).
| HLA ligand | Total | T2D | CTR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T2D, (N = 110) | Controls, (N = 55) | χ2 | Pχ | HHV8(+), (N = 78) | HHV8(−), (N = 32) | χ2 | Pχ | HHV8(+), (N = 8) | HHV8(−), (N = 47) | χ2 | Pχ | |
| 2DL2-2DL2/2DL2-2DL3 | ||||||||||||
| C1C1 | 53 (48) | 17 (31) | 48 (62) | 5 (15) | 3 (38) | 11 (23) | ||||||
| C1C2 | 30 (27) | 24 (44) | 5.59 | 0.018 | 20 (26) | 10 (31) | 25.2 | <0.0001 | 3 (38) | 24 (51) | 0.79 | 0.37 |
| C2C2 | 27 (25) | 14 (25) | 10 (12) | 17 (53) | 2 (26) | 12 (26) | ||||||
| HLA ligand | Total | T2D | CTR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T2D, (N = 58) | Controls, (N = 53) | χ2 | Pχ | HHV8(+), (N = 18) | HHV8(−), (N = 40) | χ2 | Pχ | HHV8(+), (N = 11) | HHV8(−), (N = 42) | χ2 | Pχ | |
| 2DL3-2DL3 | ||||||||||||
| C1C1 | 24 (41) | 19 (36) | 7 (39) | 17 (43) | 4 (36) | 19 (45) | ||||||
| C1C2 | 22 (38) | 25 (47) | 1.36 | 0.24 | 2 (11) | 20 (50) | 5.68 | 0.0171 | 4 (36) | 20 (48) | 3.52 | 0.06 |
| C2C2 | 12 (21) | 9 (17) | 9 (50) | 3 (7) | 3 (28) | 3 (7) | ||||||
- χ2, Chi-squared test value; Pχ, P-value obtained with Chi-squared test.
DISCUSSION
Although the primary cause of type 2 diabetes is unknown, it is hypothesized that viral infection might be one of the risk factors in addition to having a specific genetic background.
In particular, preexisting chronic infections have been associated to increased risk of developing type 2 diabetes, including infections with hepatitis C virus (HCV) [Lecube et al., 2004; Granados-Montiel et al., 2011], herpes simplex 1 (HSV-1) [Sun et al., 2005], human cytomegalovirus (CMV) [Chen et al., 2012], and HHV8 [Ingianni et al., 2007; Sobngwi et al., 2008].
On the other hand, the genetic background seems to play an equally important role in the diabetes development, and recently many reports have attempted to find correlations between the expression of specific genes and the risk to develop diabetes. Associations were observed for specific KIR and type 1 diabetes [Shastry et al., 2008; Ramos-Lopez et al., 2009; Mehers et al., 2011; Zhi et al., 2011], and post-HCV infection type 2 diabetes [Granados-Montiel et al., 2011].
The present study was therefore aimed at analyzing the possible link between HHV8 infection and KIR genes.
The study included 168 type 2 diabetes patients and 108 healthy controls, both HHV8-infected or uninfected, all belonging to the Sardinian population, who were analyzed for HHV8 presence and KIR allotype. The results showed that type 2 diabetes patients have a significantly higher incidence of HHV8 infection compared to healthy controls (57% vs. 17%) and that type 2 diabetes patients infected with HHV8 have a significantly higher frequency of KIR2DL2/DS2 genes compared to HHV8-uninfected type 2 diabetes patients, or to controls, both HHV8-infected or uninfected (81% vs. 42–51%). Even more interestingly, HHV8-positive type 2 diabetes patients showed increased homozygosity of KIR2DL2/DS2 and HLA-C1C1 genes compared to type 2 diabetes HHV8-uninfected patients (64% vs. 14%), underlining the importance of the KIR/ligand combination on immune responses and diabetes susceptibility. By contrast, none of the HHV8-positive healthy controls were homozygous for KIR2DL2/DS2 genes, suggesting a peculiar effect of KIR/HLA interaction in type 2 diabetes condition, that is not evident in healthy status.
Overall, the findings reported here suggest for the first time a possible correlation between specific KIR expression, HHV8 infection and type 2 diabetes development, indicating that, rather than a direct correlation gene/disease, HHV8 infection might be a cofactor for the development of type 2 diabetes in particularly susceptible individuals. The observed increased combination of KIR2DL2 and HLA-C1 in type 2 diabetes patients infected with HHV8, but not in HHV8-infected control subjects, suggests that KIR2DL2/HLA-C1 individuals might be more susceptible to HHV8 infection, and more prone to develop HHV8-associated alterations potentially linked to the development of diabetes, as a result of an impairment of NK cells function in controlling HHV8 infection.
This hypothesis is consistent with recent data supporting a genetic association between KIR2DL2/DS2 and susceptibility to viral infections [Estefania et al., 2007; Moraru et al., 2012], and with previous observations, showing that this specific KIR/HLA genotype is associated with reduced activity of NK cells against HSV-1 in multiple sclerosis patients [Rizzo et al., 2012]. Uncontrolled HHV8 infection might contribute to type 2 diabetes development through different mechanisms. HHV8 is known to induce insulin receptor expression on endothelial cells [Rose et al., 2007a, 2007b], and was recently observed to affect insulin and glucose uptake in infected cells [Ingianni et al., 2013].
Another potential pathway is related to the cellular stress. Endoplasmic reticulum (ER) stress can be induced by diverse stressors in the cell, including obesity [Ozcan et al., 2004] and virus infections [He, 2006], leading to the development of a consequent unfolded protein response (UPR), which can in turn suppress insulin signaling [Ozcan et al., 2004]. Cellular stress was also shown to contribute to the transition of anti-inflammatory M2 macrophages into proinflammatory M1 macrophages, which has been related to insulin resistance [Olefsky and Glass, 2010]. Consistently with this, animals deficient in XBP-1, a transcriptional factor, which modulates the ER stress response, develop insulin resistance [Ozcan et al., 2004]. In addition, the UPR is related to the induction of the activating transcription factor 4 (ATF4), a key protein in the regulation of cell response to stress, representing a converging factor of many stress-induced cellular pathways. It has been recently observed that ATF4-null transgenic mice are hypoglycemic, have a thin phenotype and an increased energy consumption [Seo et al., 2009]. ATF4 was also shown to inhibit insulin secretion and insulin sensitivity in various tissues [Yoshizawa et al., 2009], whereas the absence of ATF4 significantly decreases diabetes induced by diet, as well as the consequent hyperlipidemia and hepatosteatosis, suggesting that ATF4 might be involved in glucose metabolism [Seo et al., 2009].
It should be noted that HHV8 infection can potently induce ATF4 [Caselli et al., 2012], likely as a consequence of induced ER stress and UPR, thus an uncontrolled HHV8 infection might contribute to the development of insulin resistance also by this ATF4 induction, as summarized in Figure 2.
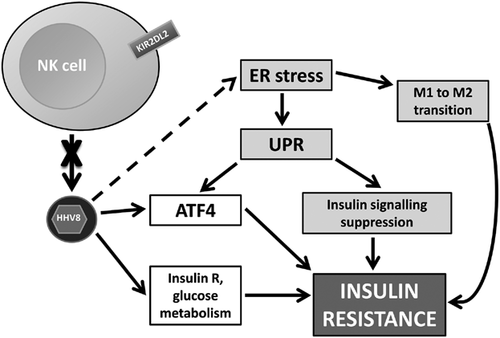
The hypothesis of a link between innate immune system, infections, and insulin sensitivity is also supported by recent observations showing that high exposure to persistent pathogens in mice is correlated to high levels of inflammatory markers and low insulin sensitivity [Krishnapuram et al., 2011].
Functional experiments are warranted in order to definitively clarify the role of KIR–HLA interactions and HHV8 infection in the pathogenesis of type 2 diabetes, similarly to what demonstrated for HSV-1 in multiple sclerosis [Rizzo et al., 2012]. Also, it might be interesting to analyze KIR2DL2 polymorphism, since recent data suggest that rs2756923 polymorphism within the KIR gene cluster haplotype B in exon 8 of the KIR2DL2 gene might have a potential role in type 1 diabetes development due to its inhibition of NK-cell cytotoxicity [Ramos-Lopez et al., 2009].
Overall, these data indicate preliminary evidence that receptors KIR2DL2/DS2 might be correlated with the presence of HHV8 infection in type 2 diabetes and suggest the possibility that such patients might have an impaired NK component contributing to the development of type 2 diabetes.
ACKNOWLEDGMENTS
We thank Daria Bortolotti for her excellent technical assistance and Linda Sartor for revising the manuscript.




