Clonal analysis of hepatitis B viruses among blood donors from Joinville, Brazil: Evidence of dual infections, intragenotype recombination and markers of risk for hepatocellular carcinoma†
Esmeralda A. Soares and Cibele R. Bonvicino contributed equally to this work.
Abstract
Hepatitis B virus (HBV) is classified into seven major genotypes (A–H). Brazil, a country of continental proportions, has three prevailing lineages of HBV genotypes A, D, and F. Distinct HBV genotypes have been associated with differential risk of disease progression. Pre-S gene deletions and single nucleotide polymorphisms have also been linked to progression to liver diseases. In this study, the molecular epidemiology of HBV was examined in Southern Brazil. The occurrence of multiple HBV infections, HBV recombination, and genetic markers of disease progression were also evaluated. Seventy-eight persons infected with HBV had their viruses characterized molecularly by nested PCR, DNA sequencing, and phylogenetic inference. Multiple infections and recombinant viruses were evaluated by clonal and bootscanning analyses. The vast majority (96%) of the strains belonged to different D subgenotypes. Three of the four strains with unresolved genotypic classification showed evidence of dual infections with distinct D subgenotypes by clonal analysis. There was also evidence of intragenotype mosaic viruses. While four viruses had pre-S deletions as major variants, another two displayed minor variants with such characteristics. One strain carried the F141L mutation, associated recently with increased risk of hepatocellular carcinoma. These results emphasize the need for monitoring HBV genotype distribution around South America, as well as for the presence of genetic markers of disease progression in subjects diagnosed with HBV recently. J. Med. Virol. 83:2103–2112, 2011. © 2011 Wiley Periodicals, Inc.
INTRODUCTION
Infection by hepatitis B virus (HBV) is a major health problem, representing the leading cause of liver cancer worldwide [Shepard et al., 2006]. More than 2 billion people have been infected to date, and between 350 and 400 million are currently HBV chronic carriers with an increased risk of developing cirrhosis and hepatocellular carcinoma (HCC) [Deny and Zoulim, 2010]. HBV-related liver disease and HCC account for approximately 1 million deaths per year [Schutte et al., 2009].
HBV is currently classified in eight genotypes (A–H) [Kurbanov et al., 2010] in addition to two other ones (I and J) which are novel and controversial [Kurbanov et al., 2008; Tatematsu et al., 2009]. Genotypes can be classified further in subgenotypes by phylogenetic clustering. HBV genotypes and subgenotypes are heterogeneously distributed around the world; while genotypes A and D are predominant in Europe, genotypes B and C are found more frequently in South and Southeast Asia and the Pacific. In South America, genotypes A, D, and F are predominant, while in North America, the majority of genotypes have been identified [Kurbanov et al., 2010].
Brazil shows a low to intermediate HBV endemicity, with a 0.37% prevalence of HBsAg, with differences between states with a slightly higher prevalence in the South, like Santa Catarina state [Brasil, 2011a]. HBsAg prevalence in blood banks has been found to be higher than in the general population and as high as 0.6% in some cities of the Southeastern region. Brazilian blood banks carry out compulsory HBsAg and anti-HBc screenings [Brasil, 2011b], and anti-HBc reactivity rates have been found to range between 5% and 11% [Brasil, 2011a]. Brazil shows similar HBV prevalent genotypic patterns to other South American countries, with highly frequent A, D, and F genotypes [Mello et al., 2007] but with significant regional disparities. While genotype A is the most prevalent in several Brazilian states, including those in the Amazon region where the HBV prevalence is highest, genotype D is predominant in Southern Brazil [Mello et al., 2007]. Several recent reports on HBV genotypes in Brazil [Gomes-Gouvea et al., 2009; Becker et al., 2010; Haddad et al., 2010; Santos et al., 2010] are restricted to specific regions (concentrated in capital cities) and have rarely investigated the presence of multiple HBV viral strains by genetic cloning.
Although data on the association between HBV genotype and clinical findings are still scarce [reviewed in Lin and Kao, 2011], there is evidence that individuals infected with the D genotype progress more frequently to chronicity and are also more likely to develop more severe liver diseases, including HCC respective to those infected with the A genotype [Sanchez-Tapias et al., 2002; Thakur et al., 2002]. Thus, HBV genotyping performed in chronically infected individuals might be helpful for identifying risk of disease progression, mainly in the Southern region of Brazil where genotype D is predominant.
In addition to HBV genotype, other viral genetic markers have been associated recently with a more severe or rapid clinical outcome, like the single nucleotide polymorphisms (SNPs) A1762T/G1764A in the viral basic core promoter, more frequently present in genotype C, associated with a higher risk of HCC [Yang et al., 2008]. These SNPs have also been reported more frequently in genotype D than in genotype A [Sharma et al., 2010]. More recently, the T55C SNP, resulting in the amino acid substitution F141L in the viral pre-S2 protein, has been associated with high risk of HCC in individuals infected with genotype C viruses [Mun et al., 2011]. Moreover, deletions in the pre-S region, including pre-S1 and pre-S2, have also been associated with more progressive liver cell damage [Fan et al., 2001; Sugauchi et al., 2003a] and as independent risk factors of disease progression and HCC [Lin et al., 2007a]. In Brazil, HBV pre-S deletions have also been reported in a single genotype A-infected blood donor [Araujo et al., 2004].
This report shows the distribution of HBV genotypes in a group of anonymous blood donors from Joinville, Southern Brazil, diagnosed with HBV infection by routine screening. These findings were carried out with aliquots of serum samples originally collected for diagnosis. In addition to describing genotype distribution, several dually infected individuals with different D subgenotypes were identified for the first time in Brazil as well as presence of intragenotype recombinants. Pre-S deletion mutants and one F141L mutant were detected in viral quasispecies in some individuals. These findings showed the relevance of conducting clonal analysis in individuals infected with HBV for a better assessment of viral diversity and its putative impact on HBV-related liver disease.
METHODS
Samples
Seventy-eight aliquots of serum samples used for HBV diagnosis, collected from individuals above 18 years of age were obtained from a regional blood bank in Joinville, in the state of Santa Catarina, Southern Brazil. In this blood bank, the annual incidence of HBV infection was approximately 5%, including anti-HBc reactivity, and repeat donors represented approximately 45% (Dr. Ozenilda Carvalho, personal communication). HBV diagnosis, carried out between 2005 and 2007, showed that all donors studied were positive for HBsAg and anti-HBc. This study was approved by the Ethical Committee of the Instituto Nacional de Câncer (INCA) in Rio de Janeiro under protocol No. 104/05 of 2/9/06.
DNA Extraction, PCR Amplification and Sequencing
Serum samples were sent to the Genetics Program of INCA for molecular analyses. Viral DNA extraction was performed with the QIAamp® DNA Mini Kit (QIAGEN, Chatsworth, CA), following the manufacturer's specifications. DNA was kept at −20°C until further use. Nested PCR was used for amplifying a fragment of approximately 1.4 kb spanning the entire pre-S/S region and part of the polymerase region (nucleotides 2,338 to 1,085 relative to the circular genomic DNA of reference strain HPBAWY—GenBank accession J02203.1). First round primers LMWRF1 (5′-CACACTCTATGGAAGGCGGG-3′) and LMWRR1 (5′-GGCCCATGACCAAGCCCC-3′), and second round PS8 and HS5-2 primers [described in Takahashi et al., 1998] were used. For the first round, reactions were conducted with 2.5 µl of 10× Taq buffer, 1 µl 1.5 mM MgCl2, 0.2 µl of 0.15 mM dNTP mix, 25 pmol of each primer, 0.25 µl of Taq Platinum DNA polymerase (Life Technologies, Carlsbad, CA) and H2O q.s.p. to 25 µl. Cycling conditions were: 94°C for 5 min, followed by 35 cycles of 94°C for 40 sec, 57°C for 30 sec, and 72°C for 1.5 min, with a final step of 72°C for 7 min for completing unfinished DNA strands. The second round was performed under the same conditions except for annealing at 50°C. PCR products were purified and sequenced with an automated 3130XL Genetic Analyzer (Life Technologies) according to the manufacturer's specifications. Sequencing reactions were performed with second round primers and additional internal primers WMFOW1 (5′-CAGGATTCCTAGGACCCCTTCTC-3′ corresponding to genomic coordinates 173 to 195 relative to HPBAWY) and WMREV1 (5′-CTTGGCCCCCAGAACCACATCATC-3′; corresponding to genomic coordinates 744 to 767). Sequences were assembled, visually inspected and edited with SeqMan (DNAStar Inc., Madison, WI) and exported as FASTA files. HBV sequences were deposited in GenBank (accession numbers JF815606–JF815693).
HBV Genotyping and Phylogenetic Analyses
HBV sequences were aligned with reference strains retrieved from GenBank representing all HBV genotypes (A–H) and their respective subgenotypes using ClustalW contained in BioEdit v.7.0.9.0 [Hall, 1999]. The best substitution model for inferring phylogenetic relationships was determined with Model Generator [Keane et al., 2006]. PhyML v.3.0 [Guindon et al., 2010] was subsequently used for maximum likelihood (ML) analysis with the previously selected substitution model (GTR + I + G). Clade robustness was evaluated by the approximate likelihood ratio test (aLRT) [Anisimova and Gascuel, 2006] and generated trees were visualized using FigTree v.1.3.1 (publicly available at http://tree.bio.ed.ac.uk/). HBV isolates were genotyped and/or subgenotyped by clustering with reference sequences.
Clonal Analyses
HBV bulk sequences showing signs of double chromatogram peaks or discordant sequences during assembly, indicating multiple strains, were analyzed by clonal analyses. Newly amplified PCR products were cloned in pCRII-Topo by T/A overhang (Life Technologies) according to the manufacturer's instructions and recombinant plasmids were transformed in DH5α chemically competent E. coli (Life Technologies). Recombinant plasmids were checked for the presence of inserts by colony PCR with PS8 and HS5-2 primers. Eleven to 16 positive clones were sequenced.
Analysis of Recombination
HBV sequences belonging to genotype D which did not group robustly with reference HBV-D subgenotypes were studied in further detail by recombination analysis. Query sequences were first aligned with representative HBV genotypes, subsequently to representatives of all D subgenotypes and subjected to bootscanning analysis with Simplot v.3.5.1 [Lole et al., 1999]. The following parameters were used: window size 200 bp, sliding step 20 bp, T/t ratio 2.0, gapstrip on, 100 replicates, Kimura-2p, neighbor-joining.
HBV Genotyping for Primary Drug Resistance
Since viral PCR products analyzed in this study contained a large part of the P gene encoding the viral polymerase, the Stanford University HBV drug resistance database (http://hivdb.stanford.edu/HBV/HBVseq/development/HBVseq.html) was used for inferring primary drug resistance.
RESULTS
HBV Genotyping and Subgenotyping
Fifty-eight HBsAg positive serum samples were PCR-amplified successfully and sequenced. Fifty-six (96%) were assigned to genotype D, while each A and F genotype was represented by a single isolate (2%). Genotyping was inferred by phylogenetic reconstructions with reference strains for all HBV genotypes as depicted in Supplementary Figure 1. Sequences belonging to genotype D were classified further into subgenotypes D1 (n = 2), D2 (n = 9), D3 (n = 34), D4 (n = 1), and D6 (n = 6). Four isolates (HBV19, HBV34, HBV39, and HBV43) could not be assigned to any subgenotype because they did not group within any genotype D cluster (Supplementary Fig. 1A). The genotype A isolate corresponded to subgenotype A1, and the genotype F isolate to subgenotype F2 (Supplementary Fig. 1B,C).
Dual HBV Infections With Different Subgenotypes
The bulk sequences of the four genotype D viruses for which subgenotyping could not be carried out consistently showed several nucleotide sites with double peaks when editing electropherograms. In addition, as different DNA strands were also observed during assembly, these HBV isolates were analyzed by cloning. PCR products of three query isolates (HBV34, HBV39, and HBV43) were newly amplified and cloned, and 11–16 individual clones from each isolate were sequenced. Interestingly, clonal analysis of these three quasispecies showed evidence of infection with two different D viruses. In all cases, mixtures of D2 and D3/D6 subgenotypes were observed (Fig. 1A–C). One D2 and 13 D3/D6 clones were identified in HBV34, 5 D2 and 11 D3/D6 in HBV39, and 6 D2 and 5 D3/D6 in HBV43. Of note, reference and query sequences belonging to subgenotypes D3 and/or D6 could not be separated unequivocally by phylogenetic analyses, raising the possibility that our query isolates could be subgenotype recombinants with phylogenetic signals corresponding to both subgenotypes.
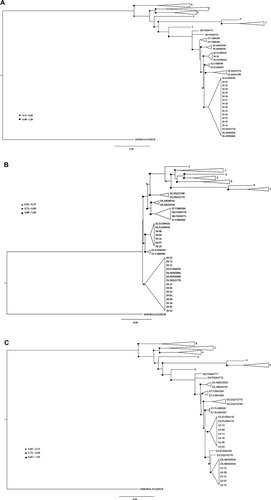
Maximum likelihood phylogenetic inference of HBV sequences generated by clonal analysis of samples HBV34 (A), HBV39 (B), and HBV43 (C). The HBV viral sequence isolated from the neotropical wooly monkey (GenBank acc. no. AY226578) was used to root all trees. Adjusted likelihood ratio test (aLRT) values are evidenced at three intervals according to the symbol legends (triangles, close circles or squares). Genetic distances between sequences are scaled by the horizontal bars at the bottom of the trees.
Evidence of Intragenotypic Recombination
Individual clones of HBV34, HBV39, and HBV43 showing dubious grouping with subgenotypes D3 and D6 were analyzed further by bootscanning. This analysis revealed the existence of diverse intragenotypic recombination patterns, involving subgenotypes D2, D3/D6, and D8. In all instances, subgenotypes D3 and D6 could not be distinguished from one another, probably due to the low number of phylogenetically informative sites in the fragment herein analyzed. Although different patterns of D3/D6 recombination were observed in clones of these three isolates (data not shown), no further attempts to classify these lineages were conducted because their bootscanning patterns varied meaninglessly due to lack of phylogenetic informative sites. On the other hand, in two isolates (HBV34 and HBV39), other subgenotype D fragments were represented. Four clones of isolate HBV34 showed pattern D3/D6_D8_D3/D6 (Fig. 2A) while two clones of HBV39 showed a D3/D6_D2 pattern (Fig. 2B). Patterns of recombination were unique and consequently not shared by different isolates, suggesting that recombinants have been most likely generated in the individuals herein studied rather than representing recombinant forms circulating in the population.
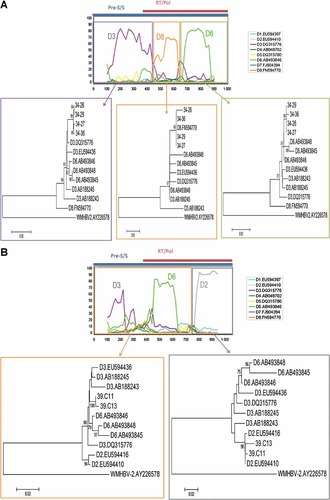
Bootscanning and subfragment phylogenentic analyses of representative clonal sequences of strains HBV34 (A) and HBV39 (B). At the top of each Figure, the bootscanning analysis to classify D subgenotypes is shown, by comparing each query sequence against reference sequences of all D subgenotypes available retrieved from GenBank. Bootscanning running parameters were as stated in the “Methods” Section. At the top of the bootscanning, the corresponding HBV genomic regions of RT/pol and pre-S/S are shown. Below each subfragment delimited by colored boxes, the phylogenetic inference of the query sequences is shown. Only bootstrap values above 75% are shown. Genetic distances between sequences are scaled by the horizontal bars at the bottom of the trees.
Pre-S/S Deletions and Point Mutations in Majority and Minority HBV Populations
HBV genotyping revealed in-frame deletions of different size in the pre-S/S coding region of four bulk sequences (HBV08, HBV27, HBV46, and HBV59). While HBV08 and HBV27 displayed small deletions, of codon 109 and codons 107–108, respectively, HBV46 and HBV59 showed more extensive deletions, accounting for 78 and 39 nt, resulting in loss of codons 106–131 and 52–64, respectively (data not shown). Except for the last deletion, all others occurred at the vicinity of, or at the start codon of the pre-S2 protein.
Clonal analyses of isolates HBV34 and HBV39 showed in-frame deletions affecting the pre-S1 coding region which were unnoticed by standard bulk sequencing due to their low frequency in each viral quasispecies. Analyses of HBV34 clones showed two types of deletion: 3/14 with loss of 51 nt (codons 50–66) and 1/14 with loss of 18 nt (codons 61–66) (Fig. 3A). Among HBV39 clones, 3/9 showed a 183 nt deletion (codons 47–107) and 1/9 a 9 nt deletion (codons 108–110) (Fig. 3B). Here again, HBV39 deletions involved the surroundings of the pre-S2 start codon.
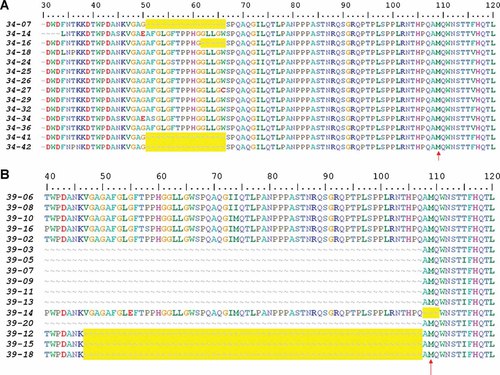
Amino acid alignment of HBV clonal sequences of strains HBV34 (A) and HBV39 (B). The amino acid numbering above the alignments correspond to the codon positions of pre-S/S protein. Dashes in the alignments represent gaps in sequences, and are highlighted in yellow. The vertical arrows indicate the starting codon of the pre-S2 protein.
The F141L point mutation in pre-S/S, recently described as a marker for increased risk of HCC in individuals with genotype C infection [Mun et al., 2011], was observed in one isolate belonging to subgenotype D2 (HBV14). Another isolate (HBV03) displayed mutation P120S, associated with vaccine escape [Oon et al., 1999].
Primary Antiviral Drug Resistance
The HBV genomic fragments examined allowed the assessment of primary antiviral drug resistance. Upon submission of sequences to the Stanford University HBV drug resistance database no primary mutations were observed. Three isolates (HBV37, HBV40, and HBV48) showed mutation I233V, whose resistance to adefovir is controversial [Schildgen et al., 2006; Curtis et al., 2007], a reason why this mutation is not included currently in HBV genotyping algorithms.
DISCUSSION
Analysis of HBV infected individuals from Joinville, southern Brazil, showed a prevalence of 96% of genotype D, 2% of genotype A, and 2% of genotype F. This observation was consistent with previous molecular studies of HBV in blood donors from Brazil [Araujo et al., 2004; Tonetto et al., 2009]. The vast predominance of genotype D herein observed was also in agreement with studies in the southern region of Brazil, where the present study was conducted [Carrilho et al., 2004; Becker et al., 2010; Bertolini et al., 2010]. It has been postulated that HBV-D is highly prevalent in southern Brazil due to the prevalence of this genotype in Central Europe, a source of intense influx of immigrants to Southern Brazil in the early 1900s [Mello et al., 2007]. HBV-D was represented by several subgenotypes (D1, D2, D3, D4, and D6), which are also common in European countries. Very few studies in Brazil showed subgenotypic data, all restricted to the Amazonian (Northern) region [Victoria Fda et al., 2008; Gomes-Gouvea et al., 2009; Santos et al., 2010], where genotype D is not the most prevalent and subgenotypes A1, F2, D1, D3, and D4 were also identified. The present study reports, for the first time, HBV subgenotypes in southern Brazil, a region of epidemiological relevance [Ministério da Saúde do Brasil, 2010].
Among the 58 HBV positive isolates herein analyzed, four (7%) had evidence of dual infections, which were proved by clonal analysis in three of them. To the best of our knowledge, this is the first description of dual HBV infections in Brazil. Most reports describe dual infections by different HBV genotypes, like B and C [Jutavijittum et al., 2006; Lin et al., 2007b], A and E [Olinger et al., 2006] or A and D [Trinks et al., 2008] rather than between different subgenotypes. Infection by two different subgenotypes of the same genotype should be common in areas where predominance of a single genotype occurs, like southern Brazil. Indeed, the occurrence of dual infections by D3/D6 and D2 were consistent with their prevalence, in view that these three D subgenotypes were the most frequently observed in the study group.
In addition to dual infections, clonal analyses showed evidence of recombination, for the first time reported in Brazil. Further bootscanning analyses confirmed occurrence of recombinant strains in two isolates (HBV39 and HBV34), between subgenotypes D3/D6 and D2 in the former and D3/D6 and D8 in the latter. Phylogenetic analyses of regions delimited by recombination breakpoints corroborated their classification with bootstrap robustness. Genomic recombination is a common feature of HBV, and some well characterized subgenotypes of epidemiological relevance are made up by strains of different genotypes like subgenotype B2, that harbors a small fragment of genotype C within a genotype B background [Sugauchi et al., 2002, 2003b, 2004], and the described recently subgenotype D8, carrying a fragment of genotype E [Abdou Chekaraou et al., 2010]. While intragenotype recombination has been reported less frequently than intergenotype recombination, it should be more likely to occur due to the higher homology of parental strains involved in the recombination event. This would make the identification of intragenotype recombination difficult. However, a comprehensive analysis of all full-length HBV sequences available at public databases showed a large number of intragenotype recombinants, particularly within genotypes A, D, and F/H [Simmonds and Midgley, 2005]. The D subgenotypes involved in recombination in our isolates were the most prevalent in the area of study, except for D8. The D8 region present in the recombinant D3/D6_D8_D3/D6 virus was of genotype D origin and lacked any region of genotype E. As subgenotype D8 has been described in Niger [Abdou Chekaraou et al., 2010], a country with no evident economic or cultural interaction with Brazil, it may be likely that subgenotype D8 is more ancient and/or disseminated worldwide than currently appreciated and could have already contributed to the genetic background of genotype D strains currently circulating in several continents.
Genetic characteristics of HBV have been associated with varying disease outcomes in infected individuals. The genotype itself is one factor, since genotype D has been associated with increased chronicity and liver disease when compared to genotype A [Sanchez-Tapias et al., 2002; Thakur et al., 2002]. The vast predominance of genotype D in a group of individuals with HBV is a matter of concern. In addition to genotype, other HBV genetic markers have been shown to affect the disease outcome, such as SNPs in promoters and coding regions [Yang et al., 2008; Mun et al., 2011] and deletions in the pre-S region associated with more advanced liver cell damage [Fan et al., 2001; Sugauchi et al., 2003a]. The pre-S2 mutation F141L, associated with increased risk of HCC [Mun et al., 2011], was identified in the virus of one individual studied, while pre-S deletions were found as a major strain in four individuals and as a minor variant in another two of the studied group. Pre-S deletions were thus observed in 6/58 (10%) isolates studied, representing an unfavorable prevalence. The fact that pre-S deletion minor variants were detected in two of three viral quasispecies analyzed by cloning suggests that the prevalence of infected individuals carrying deletion mutants is likely to be underestimated in population screening. Detailed studies of viral quasispecies should be performed for evaluating the prevalence of these mutants in infected individuals. Additionally, longitudinal surveys should be carried out to assess the selective advantage and dominance of pre-S deletion mutants over time and their correlation with an unfavorable clinical outcome.
Access to anti-HBV antiviral therapy is increasing rapidly in Brazil and the prevalence of transmitted drug resistance (TDR) among drug-naïve individuals is of increasing concern [Cuestas et al., 2010; Xu et al., 2010; Bottecchia et al., 2011]. In the present study, strains carrying TDR were not found, in coincidence with the limited use and diversity of antivirals against HBV in the country. However, continued surveillance is required to warrant early detection of TDR and public health care action in a scenario of expanding access to anti-HBV drugs in Brazil.
In summary, evidence of dual HBV infections and intragenotype recombination of viral strains has been shown for the first time in Brazil, identified in aliquots of the original samples used in diagnosis. The high frequency of genotype D in Southern Brazil, associated with the high prevalence of viruses harboring genetic markers of unfavorable clinical prognosis require continuous monitoring of the molecular epidemiology of HBV viruses in the area and in neighboring countries of South America.
Acknowledgements
We thank Prof. Héctor N. Seuánez (Instituto Nacional de Câncer—INCA—and Universidade Federal do Rio de Janeiro) for the critical reading of this manuscript and for providing the infrastructure necessary to the conduction of this study. The study was part of the requirements for LMWR to obtain her Master's of Sciences degree in Oncology (Genetics) from the Graduate Program in Oncology at INCA.




