Genetic diversity of echovirus 30 involved in aseptic meningitis cases in Brazil (1998–2008)†‡
Virus nomenclature: Order Picornavirales, Family Picornaviridae, Genus Enterovirus, Human Enterovirus B, Echovirus serotype 30.
The authors declare that there are no conflicts of interest.
Abstract
Aseptic meningitis is one of the most common neurological disorders caused by enteroviruses. Among them, Echovirus 30 (E30) is described as the main etiological agent of many outbreaks and sporadic cases. This study investigated the genomic variability of E30 isolated from the cerebrospinal fluid (CSF) of aseptic meningitis cases that occurred from 1998 to 2008 in Brazil. Over a 10-year period (1998–2008), 302 non-polio enteroviruses were isolated, of which 177 were identified as E30 (58.6%). Phylogenetic analysis of the complete VP1 gene (876 nt) of 48 E30 isolates was performed and compared with additional Brazilian and foreign strains. E30 VP1 sequences segregated into three distinct major groups and seven subgroups, which were linked to the isolation year. In general, sequence divergence among E30 strains ranged from 0.2% to 13.8%. A common direct ancestor for this set of E30 strains was not defined. Brazilian isolates from Group I were related genetically to a 1997 USA isolate and both may have a common origin. Group III representatives showed close relationship to the 2007 Argentinean isolates. The present results complement existing data on the molecular characterization and genetic variability of E30 and may contribute to the understanding of the epidemiology of aseptic meningitis in the region. J. Med. Virol. 83:2164–2171, 2011. © 2011 Wiley Periodicals, Inc.
INTRODUCTION
Aseptic meningitis, acute flaccid paralysis, and encephalitis are clinical disorders caused by enteroviruses in humans. Although most cases are asymptomatic, high morbidity rates present primarily in children and severe infections may result in serious sequels [Pallansch and Roos, 2001].
Enteroviruses consist of small, positive-stranded RNA viruses belonging to the family Picornaviridae. Based on the genetic characteristics of the VP1 capsid gene, these viruses have been classified as human enterovirus species HEV-A, HEV-B, HEV-C, and HEV-D [Stanway et al., 2005].
Many outbreaks of aseptic meningitis have been described in several countries with enteroviruses as the etiological agents. In addition to causing outbreaks, aseptic meningitis can present as sporadic cases (SC), keeping active the circulation of the viral serotypes involved in these cases [Thoelen et al., 2003; Kmetzsch et al., 2006; Grenón et al., 2008; Mirand et al., 2008; Tavakoli et al., 2008; Papa et al., 2009]. Detection and typing of the enterovirus involved in clinical disorders is valuable in relating the serotypes to these diseases and contributes to their surveillance [Tavakoli et al., 2008].
Echovirus (Enteric Cytopathogenic Human Orphan virus) serotype 30 is a member of the HEV-B species; this species includes all other echoviruses, Coxsackie B viruses, among others. Echovirus 30 (E30) is one of the most commonly isolated echoviruses and has often been associated with aseptic meningitis, responsible for many of the outbreaks and sporadic cases worldwide throughout history [Oberste et al., 1999; Trallero et al., 2000; Savolainen et al., 2001; Bailly et al., 2002; Dos Santos et al., 2006; Cabrerizo et al., 2008; Castro et al., 2009; Hayashi et al., 2009]. Surveillance of aseptic meningitis and its etiological agents is essential, considering its epidemic potential.
To elucidate the genetic variability and relationship of E30 isolated from cerebrospinal fluid (CSF) of aseptic meningitis cases that occurred in Brazil over a period of 10 years (1998–2008), sequences of complete VP1 gene of E30 were determined and phylogenetically analyzed. These sequences were also compared with Bastianni E30 prototype strain (isolated in 1958) and with complete VP1 sequences of E30, isolated worldwide, temporally associated to the period of study.
MATERIALS AND METHODS
Patients and Clinical Specimens
The specimens used in this study were CSF of aseptic meningitis cases. From December 1998 to December 2008, 3,186 CSF specimens were received at the Enterovirus Laboratory from the following Brazilian regions: Northeast (Bahia, Pernambuco, and Piauí states), West-Central (Distrito Federal), Southeast (Espírito Santo, Minas Gerais, and Rio de Janeiro states), and South (Paraná and Rio Grande do Sul).
All experiments were performed in compliance with the relevant laws and institutional guidelines and in accordance with the ethical standards of the Declaration of Helsinki. The institutional committee CEP-IPEC/FIOCRUZ approved the experiments.
Virus Isolation
The continuous cell lineages RD and HEp2 were used for enterovirus isolation, as recommended by the World Health Organization [WHO, 2004].
Molecular Typing of Non-Polio Enterovirus (NPEV)
All isolated NPEV were identified initially by VP1 partial sequencing [Dos Santos et al., 2006], followed by sequence comparisons using the BLAST tool [Altschul et al., 1990]. Among them, 48 E30 isolates were selected for phylogenetic analysis. These isolates were chosen as representative of the outbreaks and sporadic cases that occurred from 1998 to 2008.
E30 Complete VP1 Gene Sequencing
Complete VP1 gene sequences were obtained from 48 E30 isolates. Information on these strains are shown in Table I. PCR amplification after cDNA synthesis was performed with primers 008 (GCRTGCAATGAYTTCTCWGT; nt 2411–2430) and 011 (GCICCIGAYTGITGICCRAA; nt 3408–3389), which hybridize in the VP3 and 2A genes, respectively, flanking the VP1 gene [Oberste et al., 1999].
| Isolate identification | Year of isolation | Localitya | Epidemiological origin | % of Nucleotide similarity with Bastianni strain (876 nt) | % of Amino acid similarity with Bastianni strain (292 aa) | Accession number |
|---|---|---|---|---|---|---|
| 01 | 1998 | PR | Outbreak 1 | 83.5 | 92.4 | HQ152879 |
| 02 | 1998 | PR | Outbreak 1 | 83.5 | 92.8 | HQ152880 |
| 03 | 1998 | PR | Outbreak 1 | 83.0 | 91.4 | HQ152881 |
| 04 | 1998 | PR | Outbreak 1 | 83.5 | 92.8 | HQ152882 |
| 05 | 1998 | PR | Outbreak 1 | 83.1 | 91.1 | HQ152883 |
| 06 | 1999 | PR | Sporadic case | 82.6 | 91.1 | HQ152884 |
| 07 | 2001 | PR | Outbreak 2 | 82.8 | 91.1 | HQ152885 |
| 08 | 2001 | PR | Outbreak 2 | 83.2 | 91.4 | HQ152886 |
| 09 | 2001 | PR | Outbreak 2 | 83.8 | 92.1 | HQ152887 |
| 10 | 2001 | PR | Outbreak 2 | 83.9 | 91.8 | HQ152888 |
| 11 | 2001 | PR | Sporadic case | 83.5 | 91.4 | HQ152889 |
| 12 | 2001 | PR | Outbreak 3 | 83.3 | 91.4 | HQ152890 |
| 13 | 2001 | PR | Outbreak 3 | 82.9 | 91.1 | HQ152891 |
| 14 | 2001 | PR | Outbreak 3 | 82.8 | 92.1 | HQ152892 |
| 15 | 2001 | PR | Outbreak 3 | 83.5 | 90.7 | HQ152893 |
| 16 | 2002 | RJ | Sporadic case | 82.7 | 92.4 | HQ152894 |
| 17 | 2002 | RJ | Sporadic case | 83.4 | 92.8 | HQ152895 |
| 18 | 2002 | RJ | Sporadic case | 83.0 | 92.1 | HQ152896 |
| 19 | 2002 | PE | Outbreak 4 | 82.6 | 91.4 | HQ152897 |
| 20 | 2002 | PE | Outbreak 4 | 82.8 | 91.8 | HQ152898 |
| 21 | 2002 | PE | Outbreak 4 | 82.6 | 91.8 | HQ152899 |
| 22 | 2002 | PE | Outbreak 4 | 83.0 | 92.1 | HQ152900 |
| 23 | 2002 | PE | Outbreak 4 | 82.7 | 91.8 | HQ152901 |
| 24 | 2002 | PE | Outbreak 4 | 82.8 | 92.1 | HQ152902 |
| 25 | 2003 | PR | Sporadic case | 83.0 | 91.8 | HQ152903 |
| 26 | 2005 | RJ | Outbreak 5 | 82.6 | 91.8 | HQ152904 |
| 27 | 2005 | RJ | Outbreak 5 | 82.2 | 91.1 | HQ152905 |
| 28 | 2005 | RJ | Outbreak 5 | 82.4 | 91.8 | HQ152906 |
| 29 | 2005 | RJ | Outbreak 5 | 82.5 | 91.4 | HQ152907 |
| 30 | 2005 | RJ | Outbreak 5 | 82.6 | 92.5 | HQ152908 |
| 31 | 2005 | RJ | Outbreak 5 | 82.6 | 92.1 | HQ152909 |
| 32 | 2005 | RJ | Outbreak 5 | 82.0 | 91.4 | HQ152910 |
| 33 | 2005 | RJ | Outbreak 5 | 82.4 | 91.8 | HQ152911 |
| 34 | 2005 | PR | Sporadic case | 82.1 | 91.1 | HQ152912 |
| 35 | 2006 | RS | Sporadic case | 80.8 | 89.7 | HQ152913 |
| 36 | 2006 | RS | Sporadic case | 81.3 | 90.7 | HQ152914 |
| 37 | 2006 | RS | Sporadic case | 81.8 | 91.8 | HQ152915 |
| 38 | 2007 | DF | Sporadic case | 82.0 | 90.4 | HQ152916 |
| 39 | 2007 | BA | Sporadic case | 80.8 | 90.8 | HQ152917 |
| 40 | 2007 | BA | Sporadic case | 82.5 | 92.1 | HQ152918 |
| 41 | 2008 | RJ | Sporadic case | 81.2 | 91.4 | HQ152919 |
| 42 | 2008 | RJ | Sporadic case | 81.0 | 90.4 | HQ152920 |
| 43 | 2008 | RJ | Outbreak 6 | 80.5 | 91.1 | HQ152921 |
| 44 | 2008 | RJ | Outbreak 6 | 80.7 | 90.8 | HQ152922 |
| 45 | 2008 | RJ | Outbreak 6 | 81.9 | 91.4 | HQ152923 |
| 46 | 2008 | RJ | Outbreak 6 | 81.7 | 91.1 | HQ152924 |
| 47 | 2008 | RJ | Outbreak 6 | 81.7 | 91.1 | HQ152925 |
| 48 | 2008 | RJ | Outbreak 6 | 81.2 | 91.4 | HQ152926 |
- a States: BA, Bahia; DF, Distrito Federal; PE, Pernambuco; PR, Paraná; RJ, Rio de Janeiro; RS, Rio Grande do Sul.
Cycle-sequencing reactions were performed on both strands of purified PCR products, using primers 008 and 011 separately. SeqMan software from the DNAStar package was used to generate contigs of the complete VP1 gene (876 nt consensus sequences) [Burland, 2000].
Phylogenetic Analysis
Complete E30 VP1 sequences from this study were analyzed and compared with the E30 prototype strain Bastianni (USA isolate from 1958) as well as the Frater (Scotland, 1959) and Giles (USA, 1960) strains. In addition, five Brazilian E30 isolates from Pará State [Castro et al., 2009] and 57 sequences from contemporary E30 strains isolated in different countries, among complete VP1 sequences available at the GenBank, were also included.
Nucleotide and amino acid alignments of the E30 VP1 sequences and sequence edition were obtained using the Clustal-X [Thompson et al., 1997] and Bio-Edit 7.0.9 [Hall, 1999] programs.
Phylogenetic reconstruction was obtained with the MEGA 4.0 program [Kumar et al., 2001] using the Neighbor-Joining reconstruction method [Saitou and Nei, 1987]. Genetic distances were estimated using the Kimura-Two parameter model [Kimura, 1980]. Robustness of the branches in the phylogenetic tree was statistically evaluated by 1,000 bootstrap replicates [Felsenstein, 1985].
Nucleotide Sequences Accession Number
E30 VP1 sequences determined in this study are available in GenBank Database. Accession numbers are in Table I.
RESULTS
Enterovirus Isolation and Molecular Identification
From 1998 to 2008, 302 NPEV from 3,186 CSF specimens (9.4%) originating from 6 outbreaks (Table II) and 51 sporadic cases were isolated using RD and/or HEp2 cells. Altogether, 177 isolates were identified as E30 using partial VP1 sequences (58.6% of 302 NPEV). E30 was the most prevalent serotype of enterovirus isolated from sporadic cases and outbreaks during the years comprising the present study.
| Outbreak 1 | Outbreak 2 | Outbreak 3 | Outbreak 4 | Outbreak 5 | Outbreak 6 | |
|---|---|---|---|---|---|---|
| Brazilian State | Paraná State | Paraná State | Paraná State | Pernambuco State | Rio de Janeiro State | Rio de Janeiro State |
| Probable Period | December 1998 to February 1999 | December 2000 to January 2001 | October to November 2001 | April 2002 to June 2002 | March to April 2005 | October 2008 |
| Total of analyzed specimens (CSF) | 101 | 48 | 56 | 70 | 116 | 18 |
| Total of specimens with E30 isolating (isolation rate) | 35 (34.6%) | 15 (31.2%) | 21 (37.5%) | 26 (37.1%) | 22 (18.9%) | 7 (38.8%) |
| Nucleotide identity (%) among isolates | 97.3–99.3 | 98.0–99.0 | 96.9–98.6 | 98.6–99.8 | 95.0–99.3 | 94.5–99.4 |
| Amino acid identity (%) among isolates | 95.5–99.6 | 97.9–99.6 | 96.9–98.2 | 97.6–99.6 | 96.2–100 | 96.9–99.3 |
Phylogenetic Clustering of E30 Isolates
To study the genetic diversity and possible relationships of E30 circulating in Brazil from 1998 to 2008, 48 isolates were sequenced and characterized by phylogenetic analysis of the complete VP1 (876 nt). E30 VP1 sequences segregated into three distinct major groups (I, II, and III) and seven subgroups (Ia–Ic and IIa–IId) (Fig. 1). Group I included 24 sequences of E30 isolated in both outbreaks and sporadic cases in 1998, 1999, 2001, and 2002. This group was genetically homogeneous, presenting 0.2–6.1% nucleotide divergence. Clustered in Group II were 22 E30 isolates from outbreaks and sporadic cases from 2003 to 2008, presenting 0.6–8.6% nucleotide divergence. Group III consisted of two E30 isolates from sporadic cases in 2006. Bastianni prototype strain and Frater strain clustered together.
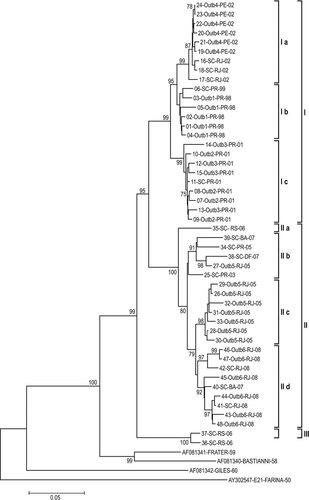
Phylogenetic analysis of E30 strains based on VP1 gene nucleotide sequences (876 nt) showing the genetic relationships between 48 Brazilian E30 strains isolated between 1998 and 2008, as well as the Bastianni prototype strain and Frater and Giles strains. The phylogenetic tree was constructed by the Neighbor-Joining method (MEGA 4.0 program). Genetic distances were estimated with the Kimura-Two parameters model. The numbers at the branching nodes are the percentage of 1,000 bootstrap replicates (percentages higher than 75% are shown). The E21 Farina prototype strain was included as an outgroup. E30 isolates were identified by number; a code differentiating outbreaks (Outbreak 1–6) from sporadic cases origin of isolate (SC); locality (see Table I for the abbreviations of the states); and the year of isolation (last two numbers of year). E30 prototype strain and Frater and Giles strain sequences were retrieved from the GenBank and identified by the accession number, name of strain and year of isolation. Major genetic groups and subgroups are indicated.
In general, nucleotide sequence divergence in pairwise comparisons among Brazilian E30 isolates ranged from 0.2% to 13.8%. Compared with the prototype strain, the genetic divergence increased to 16.1–19.5%. All Brazilian isolates characterized in this study grouped monophyletically.
The most divergent isolates were 36-SC-RS-06 and 37-SC-RS-06 (Group III), which differed from all other isolates by 10.3–13.8%. Isolate 35-SC-RS-06 did not cluster with any other isolate.
Subgroups within Groups I and II were tightly linked to the year of isolation, as a high genetic relationship among isolates of the same outbreak or sporadic cases of the same year was evident. Divergence in the E30 sequences of the 1998–1999 isolates was 0.7–3.0%. In 2001, isolates diverged 0.8–3.6%. In 2002, genetic divergences in VP1 sequences varied from 0.2% to 2.6%; in 2008, isolates diverged 0.6–5.5%.
An E30 isolate from a 1999 sporadic case (06-SC-PR-99) and all isolates from a 1998 outbreak occurred in a single geographic area (Paraná State) and clustered in a single subgroup in Group I (Ib). E30 VP1 sequences of 2001, 2002, and 2008, from both sporadic cases and outbreaks, clustered together in three respective subgroups of Groups I and II (Ic, Ia, and IId). Similar behavior was evident in E30 strains of 2005, except for isolates 27-Outbreak 5-RJ-05 and 34-SC-PR-05, which shared 3.7–5.9% nucleotide divergence when compared with other 2005 isolates.
No outbreaks were recorded in the time interval between 2003 and 2004, whereas one E30 sporadic case (25-SC-PR-03) was identified.
All 48 E30 strains were compared with additional Brazilian E30 strains isolated in 2005 and 2006 from Pará State [Castro et al., 2009]. This comparison was performed using partial sequences of the VP1 gene (349 nt). Isolate 39-SC-BA-07, a 2007 E30 strain from Bahia State sporadic case, was genetically related to the Pará E30 strains (3.2–4.6% nucleotide divergence). These strains clustered together in Group II (see Supplemental Material 1).
Comparison Between Brazilian E30 Isolates and Worldwide E30 Sequences
Phylogenetic analysis included comparisons of the VP1 sequences from the 48 Brazilian E30 isolates characterized in this study with VP1 sequences from circulating isolates during the study period (1998–2008), representative of other countries. Groups I, II, and III were compared with 46 sequences from E30 isolates collected from 1994 to 2008 (Fig. 2). Comparisons with South American isolates were performed separately, using partial E30 VP1 sequences from Argentina (420 nt).
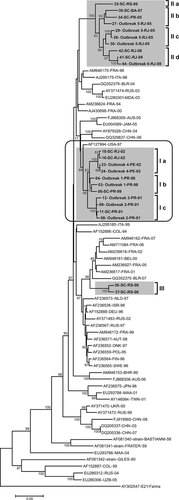
Phylogenetic analysis of E30 strains based on VP1 nucleotide sequences (876 nt) showing genetic relationships between 48 Brazilian E30 strains isolated between 1998 and 2008 (Groups I, II, and III), as well as the Bastianni prototype strain, the Frater and Giles strains and 46 isolates from the following countries: Australia (AUS), Austria (AUT), Bahrein (BHR), Belarus (BLR), Belgium (BEL), China (CHN), Colombia (COL), Denmark (DNK), Finland (FIN), France (FRA), Germany (DEU), Israel (ISR), Italy (ITA), Jamaica (JAM), Japan (JPN), Malaysia (MAA), Moldavia (MDA), Netherlands (NLD), Poland (POL), Russia (RUS), Sweden (SWE), Taiwan (TWN), Ukraine (UKR), United States of America (USA), and Uzbekistan (UZB). Phylogenetic reconstruction parameters and identification of E30 isolates were similar to those detailed in Figure 1. The E21 Farina prototype strain was included as an outgroup. E30 sequences retrieved from the GenBank were identified by the accession number, international three-letter country code and year of isolation.
When compared with E30 isolates circulating in other countries, Brazilian isolates from both Groups I and II remained as monophyletic groups. Interestingly, a 1997 USA E30 isolate [Oberste et al., 1999] was genetically related to the isolates from Group I. This E30 isolate diverged from Group I sequences 2.4–5.7%.
Analysis of the partial VP1 sequences demonstrated that two 2007 Argentinean isolates presented as closely related to isolates of Group III (5.8–6.0% genetic divergence) (see Supplemental Material 2).
Amino Acid Variability of E30 VP1 Sequences
E30 sequences obtained in this study presented at least 89.7% of similarity in VP1 amino acid sequences (292 aa) with the Bastianni prototype strain. Brazilian E30 isolates had a high degree of conservation in the VP1 amino acid sequences, with similarities ranging from 92.5% to 100% (Table I).
DISCUSSION
A molecular study of genetic variability was performed to establish associations among complete VP1 gene sequences of E30 strains isolated from the CSF of aseptic meningitis cases that occurred in Brazil between 1998 and 2008. A total of 302 NPEV were isolated in cell cultures during this period. Most of the isolates (n = 177, 58.6%) were identified as E30. These isolates were from 6 outbreaks and 51 sporadic cases that occurred in 9 Brazilian states.
The frequent presence of enteroviruses in aseptic meningitis cases may be influenced by the efficiency of transmission [Oberste et al., 1999] and dissemination of these agents in to the community. A continuous circulation of E30 in Brazil has been observed in recent years. For instance, between 1998 and 2008 several sporadic cases and at least seven aseptic meningitis outbreaks with enteroviral etiology were identified. Six of them were caused by E30 [Dos Santos et al., 2006; Kmetzsch et al., 2006].
Phylogenetic relationships inferred from comparisons of 48 E30 VP1 sequences from Brazilian isolates from 1998 to 2008 revealed the existence of two main distinct genetic groups that were temporally segregated and one group with two highly divergent sequences from sporadic cases from a same year. Sequences clustering in the two main groups were further classified within distinct subgroups. Segregation of sequences within distinct subgroups was similarly related to the time of the occurrence of the aseptic meningitis cases, clustering together isolates from outbreaks and sporadic cases from different geographic areas. These data suggest distinct introductions of E30 in the Brazilian territory causing both outbreaks and sporadic cases of aseptic meningitis.
The VP1 gene was chosen due to its high genetic variation and relevance for enteroviruses immunity. VP1 is used in molecular enteroviruses identification, as well as in phylogenetic studies [Oberste et al., 1999; Künkel and Schreier, 2000; Savolainen et al., 2001; Bailly et al., 2002, 2009; Palacios et al., 2002; Racaniello, 2007]. With the widespread application of phylogenetic analysis, it is feasible to study genetic diversity, with subdivision in subgroups or lineages and tracing genetic ties between viral strains [Page and Holmes, 1998]. Genetic relationships among E30 strains from Brazil and the Bastianni prototype strain established in this study concur with reports from other countries [Bailly et al., 2000b, 2002; Mirand et al., 2006]. Divergence in pairwise comparisons among E30 strains ranged from 0.2% to 13.8%. Similar results were described previously [Oberste et al., 1999; Caro et al., 2001; Savolainen et al., 2001].
Subgroups of Groups I and II were related to the year of isolation. This was confirmed by the low values of divergences between the isolates in 2001, 2002, and 2008. These results demonstrating high genetic similarities among E30 isolates within the same outbreak are consistent with the literature [Gjoen et al., 1996; Bailly et al., 2009]. Isolates from 1998 and 1999 clustered in a single subgroup within Group I (Ib) and may have a common ancestor.
The results suggest that a simultaneous circulation of at least two different E30 variants in Brazil occurred in 2005, given that isolate 34-SC-PR-05 did not cluster with other isolates. The geographical origin of the isolates supports the hypothesis of the co-circulating isolates in 2005: isolate 34 was from a sporadic case from the Paraná State, whereas the remaining isolates were from an outbreak that occurred in the Rio de Janeiro State. Sporadic cases are more likely to have independent origins than outbreaks.
Three strains (35-SC-RS-06, 36-SC-RS-06, and 37-SC-RS-06) from sporadic cases that occurred in the Rio Grande do Sul State in 2006 were substantially different from other E30 isolates. They appear to have been introduced independently in Brazil.
The clustering pattern of 39-SC-BA-07, an isolate from the Bahia State in 2007 and Pará E30 strains clearly suggest a recent common linking ancestor. In general, E30 isolates circulating in the same period are genetically related to each other. Group I cluster persisted for 5 years, whereas Group II persisted for 6 years.
Furthermore, based on the close genetic relationship between sequences, which presented 94–94.2% of nucleotide sequence similarity, and by the geographical proximity between the Rio Grande do Sul State and Argentina, these data also suggest that one of the E30 isolates belonging to Group III may have originated the E30 strains found in Argentina in the following year (2007). Likewise, the high genetic relationship among Group I and the 1997 USA isolate suggests a common origin.
Amino acid variability of the E30 strains demonstrated that they were highly similar to each other (92.5–100% of identity). The amino acid changes and the possible biological relevance have not been investigated. These results agree with existing E30 molecular studies [Oberste et al., 1999; Caro et al., 2001; Savolainen et al., 2001; Castro et al., 2009].
Genetic diversity in enteroviruses is relatively common and is demonstrated by the many distinct serotypes [DeFilippis and Villarreal, 2001; Stanway et al., 2005] and by variations within serotypes, as with E30 [Oberste et al., 1999; Künkel and Schreier, 2000; Savolainen et al., 2001; Bailly et al., 2002]. This serotype has been extensively studied worldwide and results corroborate its genetic variability potential [Künkel and Schreier, 2000; Bailly et al., 2000a, 2009; Savolainen et al., 2001; Palacios et al., 2002; Castro et al., 2009].
Phylogenetic analysis allowed the genetic characterization of 48 E30 VP1 sequences representative of aseptic meningitis cases from almost all Brazilian regions during a 10-year period. The knowledge acquired by this study may have significant relevance for understanding E30 behavior in terms of spread, circulation, transmission, and persistence in susceptible populations. During the progress of this study, a paper reporting the genetic relationship among five E30 isolates using partial VP1 sequences from the Pará State (Northern Brazil) was published [Castro et al., 2009]; the sequences of these five isolates were included in the analysis.
Acknowledgements
We are grateful to the Enterovirus Laboratory staff (National Reference Centre—IOC/FIOCRUZ) for the excellent technical assistance.




