Comparison of a UL111a real-time PCR and pp65 antigenemia for the detection of cytomegalovirus
Abstract
Surveillance of cytomegalovirus (CMV) replication in transplant patients is crucial for the success of transplantation. To compare a CMV pp65 antigenemia (pp65Ag) and a quantitative real-time PCR targeting the CMV-UL111a (UL111aPCR), all whole blood samples taken between July 2008 and October 2009 were identified which had been analyzed prospectively by both assays in parallel. Discordant results were re-analyzed using a published CMV duplex PCR targeting regions UL55 and UL123exon4. Of 720 samples from 81 transplant patients, CMV replication was detected in 244 specimens (34%) by the UL111aPCR (median, 1,019 geq/ml), compared to 113 (16%) detected by the pp65Ag (median, 2/250,000 leukocytes). Concordant UL111aPCR/pp65Ag results were obtained in 561 (78%) samples, being positive in 99 (14%), and negative in 462 (64%). As a rule of thumb, 1 pp65Ag-positive cell per 250,000 leukocytes corresponded to 1,000 geq/ml CMV DNA of whole blood. Discordant results were found in 159 samples (22%), being UL111aPCR-positive/pp65Ag-negative in 145 (91%; median, 650 geq/ml), or UL111aPCR-negative/pp65Ag-positive in 14 (9%; median, 1/250,000 cells). Using the duplex PCR targeting the CMV UL55 and the UL123-exon4 genes, 131 of 139 (94%) discordant UL111aPCR-positives (median UL111aPCR, 639 geq/ml; median UL55PCR, 715 geq/ml; median UL123PCR, 1,103 geq/ml) were confirmed. Of 14 discordant pp65Ag-positives, duplex PCR was also negative in 8, and of low copy number in 6. Thus, CMV UL111aPCR provides more sensitive quantitation of CMV replication than pp65Ag, however, discordant results can occur at very low viral loads. J. Med. Virol. 83:2143–2150, 2011. © 2011 Wiley Periodicals, Inc.
INTRODUCTION
Cytomegalovirus (CMV) is an important human pathogen, which is responsible for a broad range of diseases affecting immunocompromised hosts. CMV establishes a non-replicative, latent infection in CD34+ myeloid progenitor cells as a major site, [Sinclair and Sissons, 2006] and reactivates in patients with altered immune functions (for review see [Egli et al., 2008; Razonable and Eid, 2009]). Patients undergoing hematopoietic stem cell transplantation (HSCT), or solid organ transplantation are at high risk for CMV reactivation with significant direct and indirect effects causing end organ disease and limited graft survival [Helantera et al., 2010]. Depending on the type of organ transplant, immunosuppressive regimen, and serological risk constellation, the current clinical management involves antiviral prophylaxis or preemptive therapy to prevent CMV disease [Kotton, 2010]. Thus, CMV surveillance and replication dynamics of transplant patients depends on sensitive, specific, and preferably quantitative tests [Funk et al., 2007].
Detection of the CMV pp65 antigen (pp65Ag) in peripheral blood leukocytes has been used to monitor CMV replication and correlated with the risk of CMV disease [Boeckh et al., 2004; Kalpoe et al., 2004; Schroeder et al., 2004; Kalpoe et al., 2007]. However, the antigenemia assay requires same-day processing, is laborious, observer-dependent, and associated with limited sensitivity in leukopenic patients, including those after HSCT [Boeckh and Boivin, 1998; Razonable et al., 2002a]. Real-time quantitative PCR assays represent an important alternative for rapid diagnosis of CMV replication, and for monitoring the effect of antiviral therapy [Machida et al., 2000; Bordils et al., 2005]. However, the choice of plasma versus peripheral blood mononuclear cells, or whole blood is controversial as well as the identification of threshold values for initiating antiviral therapy. Overall, CMV-DNA quantitation showed a good correlation with the antigenemia assay [Gault et al., 2001; Yakushiji et al., 2002], but discordant results were also reported depending on the PCR target and the level of CMV replication [Griscelli et al., 2001; Sanghavi et al., 2008]. In the present study, an in-house real-time quantitative CMV PCR targeting the UL111a gene was compared with the pp65Ag assay using whole blood samples obtained from solid organ and hematopoietic stem cell transplant recipients. A duplex quantitative PCR [Boeckh et al., 2004] served as a reference test for discordant results.
MATERIALS AND METHODS
Clinical Specimens
Seven hundred and twenty whole blood samples from 81 patients (52 men, median age 57, range 2–73; 29 women, median age 57; range 2–75) were identified that had been analyzed in parallel for CMV by UL111aPCR and by pp65Ag, in the Division of Diagnostics of the Institute for Medical Microbiology of the University of Basel between July 2008 and October 2009. The majority of samples (88%) came from the transplant outpatient clinic of the University Hospital in Basel, Switzerland. All samples were processed within 24 hr of the blood collection.
pp65 Antigenemia Assay
The CMV pp65 antigenemia assay was used for detection of viral proteins in leukocytes as described previously [The et al., 1990; Dickenmann et al., 2001]. The assay was performed using the ARGENE CINAkit (Argene SA, Varilhes, France), and EDTA-anticoagulated whole blood. Briefly, after lysis of the erythrocytes using NH4Cl and centrifugation for 10 min at 200 × g, cell pellets were resuspended in phosphate-buffered saline to a final concentration of 2,500,000 polymorphonuclear leukocytes (PML) per ml. A volume of 100 µl of the cell suspension was deposited onto a glass slide using cytocentrifugation (7 min, 60 × g). The cells were fixed with formaldehyde and permeabilized for immunofluorescence staining with the monoclonal antibodies 1C3 + AYM-1, and anti-mouse IgG- and IgM-FITC. Quantitative results were expressed as the number of positive cells per 250,000 PML.
Nucleic Acid Extraction
DNA was extracted from 200 µl EDTA-anticoagulated whole blood using the MagNAPure LC Total Nucleic Acid Isolation kit on the MagNAPure LC Instrument™ (Roche Diagnostics, Rotkreuz, Switzerland) according to the manufacturer's instructions. DNA was eluted in 100 µl, of which 5 µl were used for each reaction. The DNA extracts were used immediately for CMV quantitation or stored at −20°C until use.
CMV Primers, Probes, and Reference Plasmids
Primers and probes for the single (UL111a) and duplex (UL55/UL123) PCR assays are listed in Table I. Reference plasmids containing the respective target sequence were used to assess sensitivity of the assays and as standards in real-time PCR. For the in-house PCR, 240 bp of the UL111a gene from CMV AD169 (GenBank: FJ527563.1) and Towne strain (GenBank: FJ616285) were amplified by PCR and cloned into the MCS of pBluescript KS-vector, (Stratagene, La Jolla, CA) and designated pCMVUL111a. The sequence of the insert was verified by DNA sequencing. For the duplex PCR two reference plasmids were used. pCMVUL55 was generated by inserting a 110 bp region of the UL55 target region into pUC57. Reference plasmid pCMVUL123-exon4 contained 116 bp of the UL123-exon4 target region cloned into pUC57 [Boeckh et al., 2004].
| Primer set | CMV target region | Amplicon size (bp) | References | Primer and Probe Sequences | |
|---|---|---|---|---|---|
| 1 | UL111a | 78 | This study | For | CCC GAC ACG CGG AAA A |
| Rev | TTC ATC GAG TAA AAC CTA CGT TGG T | ||||
| Probe | CAA TAA ACC GTA CCT ACG TGA C | ||||
| 2 | UL55 | 68 |
Boeckh et al. [2004 ] |
For | TGG GCG AGG ACA ACG AA |
| Rev | TGA GGC TGG GAA GCT GAC AT | ||||
| Probe | TGG GCA ACC ACC GCA CTG AGG | ||||
| 3 | UL123-exon 4 | 84 |
Boeckh et al. [2004 ] |
For | TCC CGC TTA TCC TCR GGT ACA |
| Rev | TGA GCC TTT CGA GGA SAT GAA | ||||
| Probe | TCT CAT ACA TGC TCT GCA TAG TTA GCC CAA TAC A | ||||
CMV DNA Quantitation Using Real-Time Quantitative PCR
Three primer and probe sets were evaluated (Table I). The routine real-time PCR targeting the CMV UL111a region (CMV IL-10 gene) resulted in a 78 bp amplicon, while the real-time CMV PCR targeting the UL55 (gB protein) and the UL123-exon4 (immediately early1, exon 4) gene resulted in a 68 and 84 bp amplicon, respectively [Boeckh et al., 2004]. The in-house PCR consisted of 300 nM of each primer, 200 nM probe, and 12.5 µl of twofold concentrated master mix (Eurogentec, Seraing, Belgium) including the Taq-polymerase, 5 mM MgCl2, dNTPs (including dUTP) and uracil-N-glycosylase. The UL111a probe was labeled at the 5′-end with 6-carboxyfluorescein (FAM) and at the 3′-end with minor groove binder (MGB) (Applied Biosystems UK, Warrington, Cheshire). The duplex PCR contained 400 nM of each primer and 100 nM of the probes. The two primer sets of the duplex PCR amplify 68 bp of UL55 CMV genomic region (gB protein) and 84 bp of UL123 region (immediately early1, exon 4). The UL55 probe was labeled with FAM at the 5′-end and 6-caboxytetramethylrhodamine (TAMRA) at the 3′-end. The UL123 probe contained the fluorophore VIC at the 5′-end, and TAMRA as a quencher at the 3′-end. Each real-time PCR was performed in 25 µl reaction volume, and using the ABI Fast Cycler 7500 Sequence Detector (Applied Biosystems, Rotkreuz, Switzerland). The PCR conditions consisted of one cycle at 50°C for 2 min and 95°C for 10 min, followed by 45 cycles at 95°C for 15 sec and 60°C for 1 min. For quantitation purposes a standard curve was generated using 100, 10,000, and 1,000,000 genome equivalents (geq) of the reference plasmids per reaction. Each sample was tested in quadruplicates, with one of the replicates being spiked with 1000 geq of the reference plasmid to ensure that negative results were not due to inhibition of the PCR assay.
Sensitivity of Real-Time PCR Assays
The reference plasmids pCMVUL111a, pCMVUL55, and pCMVUL123 were diluted using twofold dilution steps of the plasmids, covering plasmid copies from 100 to 0.39 copies per reaction. Dilutions were prepared in TE buffer containing 5 ng/ml salmon sperm DNA. Two independent runs of each concentration were tested in five replicates. The limit of detection was calculated by probit analysis using the SPSS software package version 18.0 and was 3.36 geq/PCR, corresponding to a lower detection limit of 336 geq/ml for plasmid pCMVUL111a, 5.38 geq/PCR for plasmid pCMVUL55, corresponding to 538 geq/ml and 7.41 geq/PCR of the plasmid pCMVUL123, corresponding to 741 geq/ml.
Specificity of the In-House and Duplex Quantitative PCR
To assess the specificity of the UL111a and UL55/UL123 duplex PCR [Boeckh et al., 2004], clinical samples as well as quality control samples (UK NEQAS, London UK, 2009/2010) were tested which contained more than 1 × 10E5 copies of herpes simplex type 1, Varicella-Zoster virus, Epstein-Barr virus, human herpes virus 6, and human herpes virus 8, as well as relevant positive and negative controls. Specificity for CMV was confirmed for all PCR assays (data not shown).
Statistical Analysis
The sensitivity and specificity, positive and negative predictive values were calculated using 2 × 2 tables. The number of antigen-positive cells and the level of CMV DNA were compared using Spearman's rank correlation test. P-values < 0.0001 were considered to be of statistical significance. Agreement between CMV viral loads and pp65Ag was assessed using the GraphPad Prism software as described by Bland and Altman [1986].
RESULTS
Comparison of CMV UL111aPCR Versus pp65Ag
By screening the laboratory information system for the CMV testing performed between July 2008 and October 2009, 720 whole blood samples were identified, which had been analyzed by both, CMV UL111aPCR and pp65Ag in the clinical routine. These samples originated from 81 patients, mainly after solid organ transplantation (n = 36) or HSCT (n = 40) and included both, pediatric (n = 4) and adult patients (n = 77). The vast majority of patients (88%) were followed up at the Transplant, or Hematology Outpatient Clinics at the University Hospital in Basel (Switzerland). No information was available on the CMV serostatus of donors or recipients. Table II summarizes the demographics of the patients.
| Parameter | No. of patients (%) |
|---|---|
| Total no. of patients | 81 |
| Median age (range) yr | 57 (2–75) |
| Adult (>18 yr) | 77 (95) |
| Pediatric | 4 (5) |
| Gender, male/female | 52 (64)/29 (36) |
| Type of transplantation | |
| HSCT | 40 (49) |
| Solid organ transplantation | 36 (44) |
| Kidney | 32 |
| Liver | 2 |
| Heart | 1 |
| Lung | 1 |
| Other diseases | 5 (6) |
| Breast cancer | 2 |
| CMV colitis | 1 |
| Lupus erythematodes | 1 |
| CMV primary infection | 1 |
CMV replication was detected in 244 samples (34%) from 66 patients (solid organ transplantation: 25; HSCT: 36) by UL111aPCR. The median CMV Viral load was 1,019 geq/ml (3.01 log10) and ranged from 46 geq/ml to 2.24 × 10E6 geq/ml (interquartile range [IQR] 420 to 3,821 geq/ml). By contrast, pp65Ag was positive in only 113 samples (16%) from 40 patients (15 solid organ transplantation; 24 HSCT; median, 2/250,000 cells; range, 1–115; IQR 1/250,000 to 6/250,000 cells).
Of the concordant results (561; 78%), 99 were positive (14%; median, 3,305 geq/ml; range, 99 geq/ml to 2.24 × 10E6 geq/ml) and 462 were negative with both methods (64%; Table III). Discordant results were obtained in 159 samples, 145 (91%) were UL111aPCR-positive/pp65Ag-negative having a low median CMV load of 650 geq/ml. Fourteen (9%) were UL111aPCR-negative/pp65Ag-positive with a median of 1/250,000 (Table III). Thus, the results are in accordance with an overall higher sensitivity of the UL111aPCR compared to the pp65Ag assay.
| pp65-positive | pp65-negative | Total | |
|---|---|---|---|
| CMV DNA-positive | 99 | 145 | 244 |
| CMV DNA-negative | 14 | 462 | 476 |
| Total | 113 | 607 | 720 |
For the 99 UL111aPCR- and pp65Ag-positive samples, a significant correlation was found in the non-parametric Spearman's rank test with a correlation coefficient of r = 0.5629 (P < 0.0001; Fig. 1A). To obtain a simple first estimate of the ratio between UL111aPCR and pp65Ag, CMV loads were grouped according to the pp65Ag results (Fig. 1B). The median CMV-DNA viral load was 2.8 log10 geq/ml (range, 1.7–4.8 log10 geq/ml) for group 1 (0 pp65Ag + cells), 2.9 log10 geq/ml (range, 2.0–4.0 log10 geq/ml) for group 2 (1 pp65Ag + cells), 3.4 log10 geq/ml (range, 2.2–4.5 log10 geq/ml) for group 3 (2–4 pp65Ag + cells), 3.6 log10 geq/ml (range, 2.3–4.3 log10 geq/ml) for group 4 (5–10 pp65Ag + cells), 4.0 log10 geq/ml (range, 2.8–4.4 log10 geq/ml) for group 5 (11–20 pp65Ag + cells), and 4.5 log10 geq/ml (range, 2.5–6.4 log10 geq/ml) for group 6 (>20 pp65Ag + cells). To relate the CMV loads of the UL111aPCR to the number of pp65Ag-positive cells among the 99 concordantly positive samples, the ratio of the CMV loads over the pp65Ag was calculated (Fig. 1C). Most of the samples (42%; n = 42) were found in the strata of 500–999 and of 1,000–1,499. Despite the high scatter of up to 2 orders of magnitude at either extreme, the data suggested that roughly 1 pp65Ag-positive cell per 250,000 corresponded to 1,000 geq/ml of whole blood. This is in agreement with the calculated y-intercept of the linear regression at 1,081 geq/ml (3.034 log10 geq/ml ± 0.08065) (Fig. 1A).
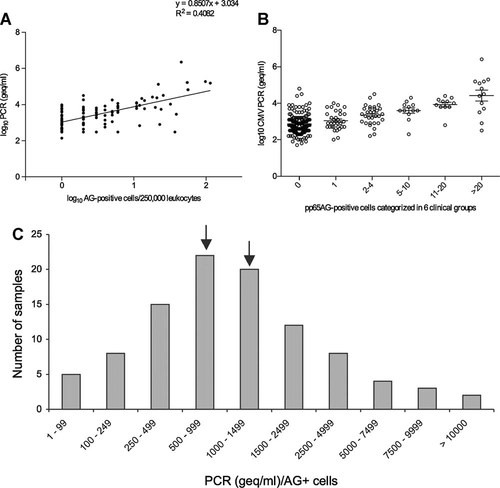
A: Correlation of CMV DNA quantitation by UL111a PCR and CMV pp65 antigenemia. The correlation was examined using non-parametric Spearman's rank test (r = 0.5629, P < 0.0001). B: Comparison of CMV-DNA geq and pp65Ag cell number. The mean CMV-DNA loads and standard error of the means are indicated. C: Ratio between PCR geq/ml and antigenemia positive cells per 250,000 leukocytes, AG: antigenemia; 99 PCR and antigenemia positive samples were used to calculate the ratio between geq/ml and antigenemia positive cells and divided into 10 groups accordingly. Indicated by arrows are the two most frequent groups, comprising 42% of the samples, showing that in this study 1 antigenemia positive cell corresponds to 500–1,500 geq/ml (in-house PCR).
Analysis of Discordant Samples Using UL55/UL123 Duplex PCR
The DNA of the 159 samples with discordant results (Table III) was re-analyzed for the presence of CMV-DNA retrospectively, using the optimized duplex PCR targeting the UL55 and UL123-exon4 genes of CMV as a reference [Boeckh et al., 2004]. Of the 145 discordant samples that were positive by UL111aPCR, but negative for pp65Ag testing, 6 could not be re-tested due to insufficient material. The UL55/UL123PCR was considered positive if one of the two target sequences was detected. Of the remaining 139 samples, the presence of CMV could be confirmed by duplex PCR in 131 (94%), and both targets were positive in 103 samples (74%). The median copy number in the 139 samples by UL111aPCR was low with 639 geq/ml (range, 46–64,980 geq/ml; IQR, 316–1,379 geq/ml). The median copy number was similarly low by the duplex CMV PCR showing 715 geq/ml for the UL55PCR (range, 7–27,660 geq/ml; IQR, 244–2,126 geq/ml), and 1,103 geq/ml for the UL123PCR (range, 21–61,173 geq/ml; IQR, 340–2715 geq/ml). The 14 UL111a PCR-negative/pp65Ag-positive samples originated from 11 patients (solid organ transplantation: 1; HSCT: 10). For 9 of the 14, a positive PCR test was available prior to the positive antigenemia testing (n = 8), or during the follow-up (n = 1). Of the five remaining pp65Ag-positive results, no preceding or subsequent samples were positive. Testing the 14 samples using the duplex UL55/UL123PCR, 8 samples (57%) were also negative. The remaining six samples were positive by UL55/UL123PCR (median, 436 geq/ml for UL55 and 197 geq/ml for UL123), but both targets being positive in only three samples. Overall, the data suggested that most of the discordant results from the pp65Ag assay resulted from PCR testing being more sensitive than pp65Ag. The contrary occurred in only a minority of cases and typically represented single events with low copy numbers.
To validate further the in-house UL111aPCR assay, results of the pp65Ag-discordant samples were compared to the UL55/UL123PCR assay using the Bland–Altman plot (Fig. 2). The results indicated a mean difference between the two tests of −0.19 log10 geq/ml (95% limits of agreement, −1.32 to 0.94 log10 geq/ml). Thus, for this subgroup, UL111aPCR and UL55/UL123PCR show a high degree of concordance.
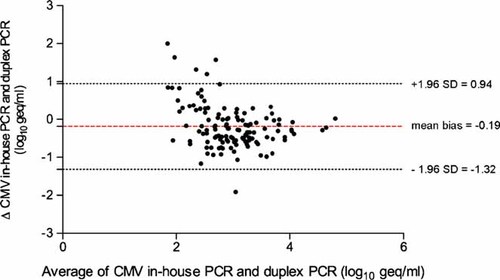
Bland–Altman plot of in-house UL111aPCR and duplex UL55/UL123PCR. Horizontal lines are drawn at the mean difference (−0.19 log10 geq/ml), and at the mean difference plus and minus 1.96 times the standard deviation of the differences (95% limit of agreement: −1.32 to 0.94 log10 geq/ml).
Monitoring of Individual Patients for CMV Viral Load
Six hematopoietic stem cell transplant patients were studied individually to elucidate the discordance between UL111aPCR and pp65Ag (Fig. 3A–F). Between 16 and 20 serial samples had been analyzed over a period of 80 (patient no. 5) to 234 (patient no. 3) days (median, 134 days) by UL111a PCR and pp65 antigenemia assay. In all six individuals, the CMV DNA results were positive, 7–94 days earlier (median, 20 days) than the first pp65-positive test and remained positive longer; for one patient (no. 4) UL111a PCR was positive during 94 days (up to 4.0 log10 geq/ml), before the first antigenemia-positive test (1 pp65-positive cell/2.5 × 10E5 cells). Furthermore, the pp65-test was intermittently negative for patient no. 3, whereas the CMV PCR remained primarily positive. CMV DNA in these six patients was always positive earlier than the pp65 antigenemia.
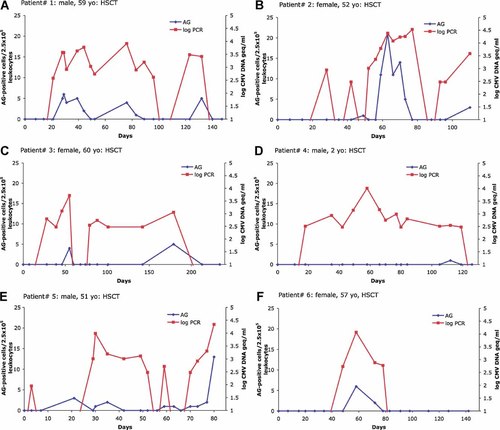
Monitoring of individual patients after HSCT for CMV load by pp65Ag or CMV UL111aPCR. On the primary y-axis the numbers of pp65-positive antigenemia cells per 250,000 leukocytes (blue diamonds) are shown, on the secondary y-axis the number of CMV-DNA log geq/ml of whole blood (red squares). yo: years old.
DISCUSSION
Quantitative assays of CMV replication have evolved as key tools for the management of patients, in terms of diagnosis and antiviral therapy, after solid organ transplantation, or HSCT. [Boeckh and Boivin, 1998]. Until recently, pp65 antigenemia has been considered the gold standard test for early diagnosis of CMV replication, but is now increasingly replaced by nucleic acid testing. In this study, the pp65 antigenemia test was compared to an established in-house real-time quantitative PCR targeting the UL111a region of the CMV genome in whole blood samples of transplant patients. The results of 720 samples from 81 patients show a high rate of concordance (78%) between UL111a PCR and pp65 antigenemia. The concordant positive samples (n = 99; 14%) showed a significant correlation with a coefficient of r = 0.5629 (P < 0.0001). For the positive samples, 1 antigenemia-positive cell per 250,000 leukocytes was found to correspond to 500–1,500 geq/ml in whole blood as measured by the UL111a PCR. Linear regression identified the y-intercept of 1,081 geq/ml. These data support that, as a rule-of-thumb, 1 pp65Ag-positive cell per 250,000 leukocytes corresponds roughly to a CMV load of 1,000 geq/ml in whole blood. A positive correlation between CMV antigenemia and CMV-DNA load has also been reported by other authors [Razonable et al., 2002b], but the details varied depending on the specimen being either plasma or whole blood, or the quantitation of CMV-DNA or mRNA [Piiparinen et al., 2001; Razonable et al., 2002a; Gerna et al., 2003; Leruez-Ville et al., 2003].
Slightly more than half of the samples, tested concordantly negative in both assays (n = 462/720; 64%). However, discordance in 22% of all samples (159/720) was discovered, the vast majority of them being UL111aPCR-positive/antigenemia-negative (n = 145). Most of these proved to be at the limit of detection, in which the UL111aPCR appeared to be more sensitive than the pp65Ag. In fact, the CMV load was below 500 geq/ml in 39% of the discordant samples. Using probit analysis, the analytical limit of detection of the UL111aPCR was 336 geq/ml. This suggested that the discordance with negative pp65Ag testing was mostly due to stochastic events at a low level of CMV replication.
The discordant UL111aPCR-positive/pp65Ag-negative specimens were reassessed with a second quantitative duplex PCR targeting the UL55 and the UL123-exon4 genes of CMV, which revealed a good correlation of in-house and duplex PCR assays (sensitivity, 96% using the duplex PCR as a reference). The Bland–Altman plot showed no significant difference in quantitation between the UL111aPCR and the duplex PCR (mean bias, −0.19 log10 geq/ml). The CMV viral load of the samples determined by the two PCR assays had low median copy numbers (639 and 715/1,103 geq/ml for the UL111a and UL55/UL123 duplex PCR, respectively), which could explain in part why pp65Ag was not detected.
Pathophysiological differences in the kinetics of DNAemia and antigenemia clearance after CMV therapy may also account for these findings [Funk et al., 2007]. Antigenemia detects CMV phosphoprotein pp65, a protein that is synthesized in excess during viral multiplication. Therefore, CMV antigen detection does not identify CMV replicating cells, but also pp65 containing cell debris phagocytosed by PML. The higher sensitivity observed with the real-time PCR analyses may provide a considerable advantage in early identification and prediction of transplant patients at high risk for the development of CMV disease, and has been described in several studies [Leruez-Ville et al., 2003; Gouarin et al., 2007; Sanghavi et al., 2008].
For a small number of only 14/720 samples, pp65Ag was positive, but the UL111aPCR tested negative. All of these specimens were in the lower range of 1–3 antigen positive cells (median, 1 pp65-positive cell/250,000 leukocytes). Similar discrepancies were observed in previous studies [Li et al., 2003; Hernando et al., 2005]. Some of the results may represent analytically false positives due to cytoplasmic background staining or reading errors, but genetic CMV variants that the PCR failed to detect cannot be ruled out. However, 8/14 samples were also negative with the UL55/UL123 PCR used as a reference. Only two of the pp65Ag-positive samples could be confirmed with the duplex PCR, but by only one of the two targets, and with very low copy number (119 geq/ml for the UL55 target sequence, 26 geq/ml for the UL123 target). The remaining represent single events suggesting that they were most likely not clinically relevant.
Previous studies have used different blood compartments for quantitation of viral loads: whole blood, plasma, peripheral blood mononuclear cells and leukocytes [Razonable et al., 2002a]. Although CMV is closely associated with leukocytes, DNA can also be detected in plasma while the virus is undergoing active viral replication. However, CMV-DNA testing on whole blood reflects both, the content of CMV in leukocytes as well as cell-free virus and has shown better sensitivity, especially when the CMV-DNA level is low [Razonable et al., 2002a]. This increased sensitivity may be of particular importance in patients after HSCT as illustrated by the time courses shown here. Consequently, at the Division of Diagnostics at the Institute for Medical Microbiology of the University of Basel, CMV loads are currently analyzed on whole blood samples for all hematological patients.
In summary, these results indicate that the quantitative real-time PCR assay for CMV UL111a is a sensitive assay to diagnose and monitor CMV replication in transplant recipients. Real-time quantitative PCR is our basis for treatment decisions, but takes clearly into account the course of the disease and the kinetics of DNA viral load more than the actual copy number.
Acknowledgements
We thank Hülya Atici and the teams of the Division of Diagnostics at the Institute for Medical Microbiology of the University of Basel for their outstanding technical support, as well as Leona Balaj-Jones for critical reading of the manuscript.




