Inverse association of IL28B genotype and liver mRNA expression of genes promoting or suppressing antiviral state†
Conflicts of Interest: None.
Abstract
High intrahepatic expression levels of interferon stimulated genes (ISGs) in chronic hepatitis C patients are associated with poor response to interferon plus ribavirin combination therapy. Expression levels of 16 genes (OAS1, PKR, MxA, ISG15, RIG-I, TLR8, IRF7, IRF9, NFKBIA, IL28A/IL28B, IL29, IL28RA, IL10RB, IFNAR2, and STAT1) that promote antiviral state and 4 genes (SOCS1, SOCS3, Zc3h12a, and A20) that suppress antiviral state were analyzed using real-time PCR assays in 133 liver biopsy samples from patients infected with genotypes 1 or 2. Expression levels of genes promoting antiviral state were positively correlated with each other but were not correlated with those that suppress antiviral state. Expression levels of some ISGs were inversely associated with common polymorphisms within the IL28B locus. Genes promoting antiviral state were expressed lower (e.g., ISG15, P = 1.42E-12 and MxA, P = 6.40E-11) in individuals with the protective rs12979860 CC genotype, and genes suppressing antiviral state were expressed higher (A20, P = 0.00107 and Zc3h12a, P = 0.00129, respectively), although some ISGs were not significant after the Bonferroni correction. The expression levels of both an antiviral (MxA) and a suppressor (SOCS1) ISG were independent predictors for non-response. These results suggest that rs12979860 genotype may be associated with response to combination therapy through an inverse relationship between antiviral and suppressor ISGs in the liver. J. Med. Virol. 83:1597–1607, 2011. © 2011 Wiley-Liss, Inc.
Abbreviations:
GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HCV, hepatitis C virus; IRRDR, interferon and ribavirin response determining region; ISDR, interferon sensitivity determining region; OAS1, 2′–5′ oligoadenylate synthetase1; PKR, double stranded RNA dependent protein kinase; SNP, single nucleotide polymorphism.
INTRODUCTION
Forty-eight weeks of peg-interferon plus ribavirin combination therapy is the current standard of care for patients infected chronically with hepatitis C virus (HCV), a positive-strand RNA virus in the family Flaviviridae [Manns et al., 2001; Hadziyannis et al., 2004], but even with this therapy the rate of sustained virological response remains 50% for genotype 1b [Adinolfi et al., 2000; Manns et al., 2001; Fried et al., 2002; Zeuzem et al., 2005]. The eradication rate may be improved slightly by extending the treatment period to 72 weeks, but this appears to be effective primarily among patients who show a rapid virological response by week 12 [Jensen et al., 2009]. Consequently, many patients relapse or fail to respond to treatment, and the high costs and severe side effects prevent some patients from receiving the full dosage and duration of therapy.
To explore why some patients are unable to eradicate the virus during combination therapy, factors affecting expression levels of intrahepatic interferon stimulated genes (ISGs) were analyzed in relation to treatment outcome. Following HCV infection, the innate immune response is triggered when intracellular RNA helicases, including retinoid acid-inducible gene I (RIG-I), detect viral dsRNA and induce production of interferon β via the adaptor protein Cardif [Kawai et al., 2005; Seth et al., 2005; Xu et al., 2005; Rehermann, 2009]. Interferons, in turn, induce a potent antiviral state by activating hundreds of ISGs through the Jak-STAT pathway [Kotenko et al., 2003]. Interferon binds to IFNAR receptors, activating the Jak1 and Tyk2 tyrosine kinases, which then phosphorylate STAT1 and STAT2, which in turn move to the nucleus and bind to the IFN-stimulated response element (ISRE) of multiple ISG promoters, thereby establishing an antiviral state within the cell and among neighboring cells [Rehermann, 2009]. A subset of suppressor ISGs, including suppressors of cytokine signaling (SOCS1) and SOCS3, negatively regulate the interferon pathway by binding to the IFN receptor and prevent STAT1 and STAT2 phosphorylation by inhibiting Jak1 and Tyk2 [Sarasin-Filipowicz et al., 2008].
HCV infection results in up-regulation of ISG expression levels in infected cells and their neighbors through activation of the innate immune response [Smith et al., 2003; Helbig et al., 2005]. Paradoxically, however, Asahina et al. [2008] recently reported that high baseline expression levels of RIG-I, ISG15, and USP18 correlate with poor response to combination therapy. Pre-treatment ISG expression levels have been found to be lower in patients who respond successfully to combination therapy than in non-responders, whereas ISGs in patients who respond poorly are often already activated at an intermediate level and cannot be further induced with interferon treatment [Sarasin-Filipowicz et al., 2008]. Conversely, interferon administration leads to a strong and rapid induction of ISGs, but the fold change was greater in responders than non-responders [Feld et al., 2007; Asahina et al., 2008], suggesting that ISG expression is either suppressed in the livers of non-responders or maintained at a higher baseline rate, preventing an effective induction [Asahina et al., 2008]. Not surprisingly, HCV has evolved multiple mechanisms to interfere with interferon production and suppress ISG induction. The effect of interferon administration on ISG expression levels diminishes following HCV infection due to viral interference, making cells less sensitive to interferon, primarily through disruption of the interferon signaling pathway [Lanford et al., 2007]. For example, the viral serine protease NS3/4A disables dsRNA sensing by cleaving and inactivating the RIG-I adaptor Cardif [Li et al., 2005]. Higher levels of endogenous interferon in non-responders may also down-regulate the RIG-I/Cardif signaling pathway, reducing signal transduction effectiveness [Asahina et al., 2008].
Although it is not clear how differences in baseline ISG expression are maintained, Honda et al. [2010] reported that expression profiles of ISGs vary by rs8099917 genotype, a single nucleotide polymorphism (SNP) upstream of the recently described type III (lambda) interferon IL28B gene on chromosome 19. rs8099917, rs12979860, and other SNPs within the same haplotype block are also significantly associated with spontaneous viral clearance, rapid and early virological response, and/or sustained virological response following treatment with peg-interferon and ribavirin for HCV genotype 1b [Ge et al., 2009; Suppiah et al., 2009; Tanaka et al., 2009]. Viral polymorphisms, including substitutions at core protein amino acid 70 and 91 and within the NS5A interferon response determining region (ISDR), are also associated with outcome of combination therapy [Enomoto et al., 1995; Enomoto et al., 1996; Akuta et al., 2006b; Akuta et al., 2007b; El-Shamy et al., 2008].
In this study, hepatic expression levels of the following interferon-related genes were analyzed: RIG-I and TLR8 (sensing of single stranded RNA); IRF7, IRF9, and NFKBIA (type 1 interferon production); IL28A/IL28B and IL29 (lambda interferons); IL28RA and IL10RB (lambda interferon receptor); IFNAR2 (type 1 interferon receptor); STAT1 (type 1 interferon signaling), 2′–5′ oligo adenylate synthetase (OAS1); double stranded RNA dependent protein kinase (PKR), MxA, and ISG15 (ISGs that induce antiviral state in cells); and SOCS1, SOCS3, Zc3h12a, and A20 (interferon signaling suppressors). It was found that expression levels of ISGs that promote antiviral state were lower in individuals with the favorable IL28B genotype, and expression levels of ISGs that suppress antiviral state were higher. The predictive value of expression levels of these genes and the contribution of clinical and viral factors on the outcome of combination therapy were investigated.
MATERIALS AND METHODS
Patients
Liver specimens were examined from 133 patients with chronic HCV genotype 1 or 2 infection who were treated at Hiroshima University Hospital between December 2002 and November 2008. Liver biopsy specimens which were obtained prior to treatment in routine clinical practice were retained at −80°C until analysis. Fibrosis stage and activity were diagnosed by pathologists at Hiroshima University Hospital as described previously [Desmet et al., 1994]. Patients received weekly injections of peg-interferon-alpha-2b for 48 weeks with dosage adjusted by body weight (60 µg for 35–45 kg, 80 µg for 46–60 kg, 100 µg for 61–75 kg, 120 µg for 76–90 kg, 150 µg for 91–120 kg). Ribavirin was administered orally with dosage based on body weight (600 mg for <60 kg, 800 mg for 60–80 kg, 1,000 mg for >80 kg). Ribavirin dosage was reduced if hemoglobin levels fell to 10.0 g/dl, and ribavirin administration was stopped if hemoglobin levels fell to 8.5 g/dl. Treatment outcome was defined as follows: Sustained virological responders were negative for HCV RNA 24 weeks after cessation of therapy; relapsers became negative for HCV RNA during the course of therapy but relapsed during the follow-up period; and non-responders never became negative for HCV RNA. Patient clinical profiles are shown in Table I. All patients provided written informed consent. The study protocol conforms to the ethical guidelines of the Declaration of Helsinki and was approved a priori by the ethical committees of Hiroshima University and RIKEN.
| Characteristic | (n = 133) |
|---|---|
| Age [median (range)] | 59 (28–80) |
| Sex (Male/Female) | 75/58 |
| ALT [median (range)] IU/L | 50 (13–543) |
| γ-GTP [median (range)] IU/L | 47 (12–708) |
| Fibrosis (F1/F2/F3/F4) | 36/43/20/33 |
| Activity (A1/A2/A3) | 33/73/26 |
| Genotype (1b/2a/2b) | 109/23/1 |
| Virus titer (log IU/ml) | 6.3 (3.6–7.4) |
| Core aa70 (wild/mutant) | 58/46 |
| Core aa91 (wild/mutant) | 50/54 |
| ISDR substitutions (0/1/≧2) | 59/21/25 |
| rs12979860 genotype (CC/CT/TT) | 84/41/8 |
| Effect of therapy (SVR/TR/NR) | 54/28/24 |
- Patients were treated with peg-interferon alpha-2b plus ribavirin combination therapy. For categorical data, the number of patients in each category is shown, and for continuous data, the median and range are displayed.
- SVR, sustained virological response; TR, transient response/relapse; NR, non-response.
Genotyping
Genotyping of IL28B SNP rs12979860 was performed using either the Invader assay or the TaqMan assay as described previously [Ohnishi et al., 2001; Suzuki et al., 2003]. For the Invader assay, allele-specific oligonucleotide pairs and invasive probes were designed and supplied by Third Wave Technologies (WI). FRET probes were labeled with FAM or VIC corresponding to alleles. The 10 µl reaction volume consisted of 0.5 µl of signal buffer, 0.5 µl of FRET probes, 0.5 µl of structure-specific cleavage enzyme, 1 µl of allele-specific probe mix, and 2 µl of PCR product diluted 1:10. Samples were incubated at 95°C for 5 min and then at 63°C for 15 min in an ABI PRISM 7700 Sequence Detector Systems (Applied Biosystems, CA), and then fluorescence data were collected. Signal intensity was calculated as the ratio of FAM or VIC to ROX, an internal reference. Genotypes were determined visually in the dye components view of the SDS software.
In the TaqMan assay, PCR was carried out using TaqMan Universal PCR Master Mix (Applied Biosystems, CA), 1 ng DNA, 0.2 µM of each primer, and 40 nM of probe provided by Applied Biosystems in 3-µl reactions. Each 384-well plate contained 376 samples of unknown genotype and 8 no-DNA control samples. Thermal cycle conditions were 50°C for 2 min, 95°C for 10 min, 50 cycles of 92°C for 15 sec, and 58°C for 1 min. Fluorescence data were collected, and genotypes were determined using the SDS software.
Quantitative Analysis of mRNA of ISGs
Total RNA was extracted from liver samples using the RNeasy Mini Kit (Qiagen, Valencia, CA). Each RNA sample was reverse transcribed with ReverseTra Ace (TOYOBO Co. Ltd., Japan) and Random Primer (Takara Bio, Kyoto, Japan). mRNA was quantified for OAS1, PKR, MxA, ISG15, SOCS1, SOCS3, Zc3h12a, and A20 with SYBR Green assay, and mRNA for RIG-I,TLR8, IRF7, IRF9, NFKBIA, IL28A/IL28B, IL29, IL28RA, IL10RB, IFNAR2, and STAT1 was quantified with TaqMan probe. IL28 gene expression includes both IL28A and IL28B expression because it was difficult to measure them separately due to high sequence similarity. Primers and probes used in this study are listed in Table II. In the SYBR Green assay, real time PCR was carried out using the power SYBR Green PCR Master Mix (Applied Biosystems, CA), 1 µl cDNA, and 400 nM of each primer in a total volume of 25 µl. Thermal cycle conditions were 50°C for 2 min, 95°C for 10 min, 45 cycles of 95°C for 15 sec, and 60°C for 1 min. In the TaqMan assay, real time PCR was performed using the TaqMan Gene Expression Master Mix (Applied Biosystems, CA), 1 µl cDNA, and 1.25 µl of 20× TaqMan probe provided by Applied Biosystems in a total volume 25 µl. Thermal cycle conditions were 50°C for 2 min, 95°C for 10 min, 45 cycles of 95°C for 15 sec, and 60°C for 1 min. Thermal cycling was done on an ABI PRISM 7300 (Applied Biosystems, CA). Results were normalized to the transcript levels of the house-keeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
| Gene symbol | Primer |
|---|---|
| GAPDH | 5′-CCACATGGCCTCCAAGGA-3′ |
| 5′-CCCAGCAGTGAGGGTCTCTCT-3′ | |
| IL28 | 5′-AAGTCCCTGTCTCCACAGGAGC-3′ |
| 5′-CTCAGCCTCCAAAGCCACG-3′ | |
| OAS1 | 5′-ACCTGGTTGTCTTCCTCAGTCC-3′ |
| 5′-GAGCCTGGACCTCAAACTTCAC-3′ | |
| PKR | 5′-TGGCCGCTAAACTTGCATATC-3′ |
| 5′-AGTTGCTTTGGGACTCACACG-3′ | |
| MxA | 5′-TTCGGCTGTTTACCAGACTCC-3′ |
| 5′-CAAAGCCTGGCAGCTCTCTAC-3′ | |
| ISG15 | 5′-TGGCGGGCAACGAATTCCAGG-3′ |
| 5′-CAGCCAGACGCTGCTGGAAGG-3′ | |
| SOCS1 | 5′-ACGAGCATCCGCGTGCACTT-3′ |
| 5′-AAGAGGCAGTCGAAGCTCTC-3′ | |
| SOCS3 | 5′-GAAGATCCCCCTGGTGTTGA-3′ |
| 5′-TTCCGACAGAGATGCTGAAGAGT-3′ | |
| Zc3h12a | 5′-GCCCCGCTCCAGAAACCAGTTC-3′ |
| 5′-CCTCAGCTCCCTCTAGTCCCGC-3′ | |
| A20 | 5′-AGGCCACCCTGGAAAGCCAGAA-3′ |
| 5′-TCTGTGTCCTGAACGCCCCACA-3′ |
| Probes | Gene symbol | Assay ID |
|---|---|---|
| DDX58 (RIG-I) | Hs00204833_m1 | |
| TLR8 | Hs00607866_mH | |
| IRF7 | Hs00185375_m1 | |
| IRF9 | Hs00196051_m1 | |
| NFKBIA | Hs00153283_m1 | |
| IL28A | Hs00820125_g1 | |
| IL29 | Hs00601677_g1 | |
| IL28RA | Hs00417120_m1 | |
| IL10RB | Hs00175123_m1 | |
| IFNAR2 | Hs01022059_m1 | |
| STAT1 | Hs01014002_m1 |
ISDR and Core Protein Amino Acid Substitutions
Amino acid substitutions in the HCV core and ISDR regions were determined by direct sequencing of PCR products following extraction and reverse transcription of serum HCV RNA. Core amino acid substitutions at positions 70 and 91 and the number of ISDR substitutions were determined as described previously [Enomoto et al., 1995; Enomoto et al., 1996; Akuta et al., 2006a; Akuta et al., 2007a].
Statistical Analysis
Data were analyzed using the Mann–Whitney U test for continuous variables and the χ2 or Fisher exact test for categorical variables using the R statistics package (http://www.r-project.org). The Bonferroni correction was used to adjust P-values for multiple testing. Univariate factors with a P-value less than 0.05 were analyzed using multiple logistic regression analysis with forward/backward stepwise selection and then validated via bootstrapping with the rms library [Moons et al., 2004].
RESULTS
mRNA Expression Levels of Interferon Related Genes
Expression levels of most of the 16 antiviral state ISGs, including RIG-I, TLR8, IRF7, IRF9, NFKBIA, IL28A/IL28B, IL29, IL28RA, IL10RB, IFNAR2, STAT1, OAS1, PKR, MxA, and ISG15 were correlated (Figs. 1 and 2A). In contrast, none of the four ISGs that suppress antiviral state (SOCS1, SOCS3, Zc3h12a, and A20) were significantly correlated with any of the genes that promote antiviral state (Fig. 2B), but they were weakly correlated with each other (Figs. 1 and 2C). Total ISG mRNA levels increased gradually with stage of fibrosis, but the ratio of GAPDH to total ISG mRNA did not differ with respect to fibrosis (P = 0.057) or other factors.
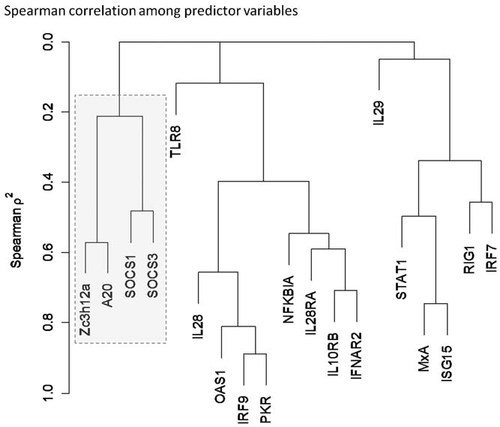
Spearman correlation among predictor variables. Hierarchical clustering identified groups of genes with similar expression patterns. Genes that suppress the antiviral state form a discrete cluster (shaded box).
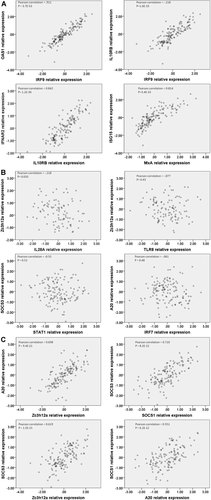
Correlation of expression levels of intrahepatic ISGs measured by real time PCR in 133 liver biopsy samples. A: Representative strong correlations among ISGs that establish anti-viral state. B: Poor correlations between expression levels of ISGs that work to establish anti-viral state and those that work to suppress interferon signaling. C: Positive correlation of expression levels among genes that work to suppress interferon signaling.
mRNA Expression Levels of Interferon Related Genes by IL28B Genotypes
Expression levels of most of the antiviral ISGs were significantly lower in patients with the rs12979860 CC genotype (Table III, Fig. 3), whereas expression levels of two out of the four suppressor ISGs (Zc3h12a and A20) were significantly higher in rs12979860 CC patients (P = 0.00129 and P = 0.00107, respectively). After the Bonferroni correction for 20 tests, the two suppressor ISGs remained significant but five of the 16 antiviral ISGs were no longer significant (Table III, Padj).
| ISG | N | CC | CT/TT | P | Padj |
|---|---|---|---|---|---|
| IL28 | 127 | 0.00016 | 0.0049 | 2.38E-05 | 0.000452*** |
| IL29 | 133 | 2.60E-05 | 0.00029 | 0.0431 | |
| RIGI | 133 | 0.05 | 0.11 | 1.44E-06 | 2.74E-05*** |
| TLR8 | 108 | 0.0019 | 0.0036 | 0.0351 | |
| IRF7 | 132 | 0.012 | 0.025 | 5.72E-05 | 0.00109** |
| IRF9 | 133 | 0.2 | 0.56 | 0.000137 | 0.00261** |
| NFKBIA | 114 | 0.075 | 0.12 | 0.0109 | |
| IL28RA | 130 | 0.0049 | 0.011 | 0.000439 | 0.00834** |
| IL10RB | 131 | 0.028 | 0.047 | 0.0318 | |
| IFNAR2 | 133 | 0.051 | 0.083 | 0.003 | |
| STAT1 | 133 | 0.17 | 0.33 | 4.32E-05 | 0.000821*** |
| OAS1 | 133 | 0.1 | 0.35 | 1.20E-07 | 2.28E-06*** |
| PKR | 132 | 0.021 | 0.3 | 3.19E-05 | 0.000606*** |
| MxA | 133 | 0.0028 | 0.017 | 6.40E-11 | 1.22E-09*** |
| ISG15 | 133 | 0.24 | 1.8 | 1.42E-12 | 2.69E-11*** |
| USP18 | 128 | 0.10 | 0.38 | 9.65E-07 | 1.93E-05*** |
| SOCS1 | 132 | 0.0021 | 0.002 | 0.884 | |
| SOCS3 | 133 | 0.055 | 0.03 | 0.0751 | |
| Zc3h12a | 132 | 0.09 | 0.038 | 0.00129 | 0.0245* |
| A20 | 132 | 0.58 | 0.29 | 0.00107 | 0.0203* |
- Median ISG expression levels and results of Mann–Whitney U test are shown by genotype for SNP rs12979860. P-values are shown for individual tests as well as after Bonferroni correction.
- Padj: Bonferroni-corrected P-values based on 20 tests.
- * P < 0.05.
- ** P < 0.01.
- *** P < 0.001.
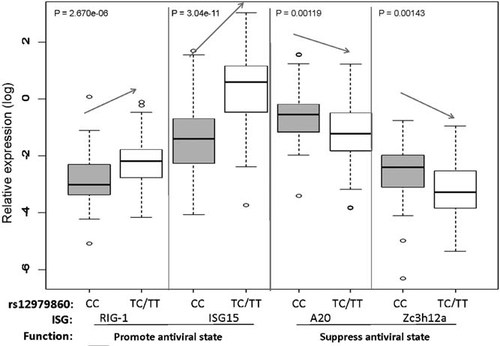
Reciprocal relationship between expression of individual antiviral and suppressor ISGs by rs12979860 genotype. Expression of two ISGs that promote antiviral defense (RIG-I and ISG15) tended to be lower in patients with the favorable rs12979860 CC genotype, whereas expression of two ISGs that suppress the antiviral state (A20 and Zc3h12a) tended to be higher in these patients. Eighty four patients had the favorable genotype (CC), and 49 patients had unfavorable genotypes (TC or TT). ISG expression levels were normalized to the transcript levels of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
IL28 Locus Genotypes and Outcome of Combination Therapy
When patients with HCV genotype 1 and 2 were tested together, the unfavorable rs12979860 genotype was significantly associated with non-response in univariate analysis (P = 0.013) but not in multivariate analysis (P = 0.116) (Table IV). When patients with genotype 1 are considered separately, rs12979860 genotype was significantly associated with outcome of therapy (sustained virological response vs. relapse or non-response: P = 0.014; non-response vs. sustained virological response or relapse: P = 0.019, data not shown), but rs12979860 genotype was not associated with outcome of treatment for patients with genotype 2 (P > 0.6).
| SVR | NVR | ||||||
|---|---|---|---|---|---|---|---|
| Simple | Multiple | Simple | Multiple | ||||
| Variable | n | P | OR | P | P | OR | P |
| Sex | 133 | 0.323 | 0.816 | ||||
| Age | 133 | 0.087 | 0.408 | ||||
| rs12979860 (CC vs. CT/TT) | 133 | 0.051 | 0.013 | 2.77 | 0.116 | ||
| Fibrosis stage | 132 | 0.020 | 0.428 | 0.052 | 0.010 | 8.76 | 0.002** |
| Activity | 132 | 0.055 | 0.466 | ||||
| ALT | 125 | 0.217 | 0.831 | ||||
| Gamma-GTP | 125 | 0.790 | 0.187 | ||||
| Core aa70 (wt vs. mutant) | 104 | 0.646 | 0.126 | ||||
| Core aa91 (wt vs. mutant) | 104 | 0.116 | 0.139 | ||||
| ISDR substitutions | 105 | 0.342 | 0.080 | ||||
| Viral load (Log IU/ml) | 128 | 0.068 | 0.018 | 2.44 | 0.040* | ||
| Genotype 1b versus 2a/2b | 133 | 0.003 | 0.48 | 0.016* | 0.148 | ||
| IL28 | 127 | 0.473 | 0.118 | ||||
| IL29 | 133 | 0.936 | 0.864 | ||||
| RIGI | 133 | 0.335 | 0.001 | 3.01 | 0.006** | ||
| TLR8 | 108 | 0.692 | 0.045 | ||||
| IRF7 | 132 | 0.130 | 0.014 | ||||
| IRF9 | 133 | 0.735 | 0.569 | ||||
| NFKBIA | 114 | 0.767 | 0.199 | ||||
| IL28RA | 130 | 0.503 | 0.328 | ||||
| IL10RB | 131 | 0.603 | 0.434 | ||||
| IFNAR2 | 133 | 0.493 | 0.961 | ||||
| STAT1 | 133 | 0.712 | 0.090 | ||||
| OAS1 | 133 | 0.598 | 0.392 | ||||
| PKR | 132 | 0.972 | 0.276 | ||||
| MxA | 133 | 0.087 | 0.023 | ||||
| ISG15 | 133 | 0.079 | 0.056 | ||||
| SOCS1 | 132 | 0.633 | 0.004 | 0.565 | 0.011* | ||
| SOCS3 | 133 | 0.827 | 0.085 | ||||
| Zc3h12a | 132 | 0.224 | 0.730 | ||||
| A20 | 132 | 0.557 | 0.359 | ||||
- * P < 0.05.
- ** P < 0.01.
Analysis of Factors Associated With Sustained Virological Response
Under univariate analysis for factors associated with sustained virological response, stage of fibrosis and HCV genotype were statistically significant at the 0.05 level. Under multiple logistic regression analysis, only HCV genotype was identified as an independent predictor for sustained virological response (Table IV). When patients with genotype 1 were analyzed separately, rs12979860 SNP genotype and expression levels of ISG15 and MxA were significant under univariate analysis, but only rs12979860 genotype was significantly associated with sustained virological response under multivariate analysis (P = 0.015, data not shown).
Analysis of Factors Associated With Non-Response to Combination Therapy
Significant univariate predictors for non-response included IL28B SNP rs12979860, stage of fibrosis, viral load, and expression levels of MxA, RIG-I, TLR8, IRF7, and SOCS1. Under multiple logistic regression analysis, only fibrosis stage, viral load, and expression levels of RIG-I and SOCS1 were significant independent predictors of non-response (Table IV). For patients with genotype 1, rs12979860 genotype, fibrosis stage, and expression levels of MxA, RIG-I, ISG15, IRF7, STAT1, and SOCS1 were significant univariate predictors of non-response, but only fibrosis stage, RIG-I, and ISG15 expression levels were significant under multivariate analysis (P = 0.011, P = 0.035, and P = 0.045, respectively; data not shown).
Effect of Viral Substitutions on ISG Expression
The effect of ISDR and amino acid 70 and 91 substitutions on the expression of individual ISGs was examined. ISG15 expression was significantly higher in patients with a substitution at amino acid 70 (P = 0.012) or 91 (P = 0.028), but expression levels of other ISGs were not associated with viral substitutions.
DISCUSSION
In this study, ISG expression levels of 20 genes involved in regulation of antiviral state via interferon signal transduction were analyzed. Positive correlations among expression levels of ISGs that promote the antiviral state were found (Figs. 1 and 2A), but expression levels of these genes were poorly correlated with expression levels of ISGs that suppress antiviral state (Figs. 1 and 2B), suggesting that positive and negative feedback systems work separately during HCV infection. However, expression levels of both promoting and suppressing ISGs were significantly associated with a common polymorphism in the IL28B locus. This result supports the assertion by Honda et al. [2010] that the stable state resulting after long-term HCV infection differs by IL28B genotype (Fig. 3).
While intrahepatic ISG expression levels have been reported to be up-regulated following HCV infection [Bigger et al., 2001; Su et al., 2002; Smith et al., 2003; Bigger et al., 2004; Helbig et al., 2005], pre-treatment ISG expression levels have been shown to be higher in non-responders than responders [Chen et al., 2005; Asahina et al., 2008; Asselah et al., 2008; Sarasin-Filipowicz et al., 2008], resulting in poor response to interferon therapy [Honda et al., 2010]. Conversely, induction of ISGs after interferon administration has been shown to be stronger in responders [Feld et al., 2007], suggesting that ISG expression in the liver is tightly regulated in order to establish an effective induction response to interferon.
Expression levels of ISGs that promote antiviral state and those that suppress it appear to be inversely related based on differences in baseline expression levels associated with IL28B SNP genotype. The favorable rs12979860 CC genotype is associated with both decreased expression of antiviral ISGs as well as increased expression of suppressor ISGs in the liver, suggesting a potential role for IL28B in intrahepatic regulation of ISG expression. The larger increase in ISG expression after IFN administration in these patients may be related to the elevated expression of genes that suppress interferon signaling, thereby facilitating a larger induction potential. While expression levels of two of the four suppressor ISGs (A20 and Zc3h12a) differ significantly by IL28B genotype following correction for multiple testing, their P-values, and effect sizes are more modest than most of the antiviral ISGs (e.g., ISG15, MxA, and RIG-I). This suggests that the association with suppression of antiviral state is relatively smaller or more indirect than the association with promotion of antiviral state, although suppressor ISG expression might also follow a different temporal pattern than antiviral ISGs. Furthermore, among the suppressor ISGs, it appears that SOCS1 expression is more directly related to treatment outcome than A20 or Zc3h12a, making it more difficult to determine the relationship between antiviral and suppressor ISGs with regard to rs12979860 genotype.
ISG expression levels also appear to vary by tissue. ISG expression profiles have been shown to differ between liver and peripheral blood cells [He et al., 2006], and several previous studies have shown that expression levels of IL28 are higher in peripheral blood mononuclear cells in patients with the rs12979860 CC genotype [Suppiah et al., 2009; Tanaka et al., 2009], whereas IL28 expression levels in the liver were lower in patients with the CC genotype.
Relative expression levels of IGS15 were significantly lower in patients with the favorable rs12979860 CC genotype [0.24 (0.11–0.50)] than in patients with CT or TT genotypes [1.8 (0.63–3.20)] (P = 1.42E-12; Table III). ISG15 has been shown to have antiviral activity against DNA and RNA viruses [Lenschow et al., 2007; Pitha-Rowe and Pitha, 2007] and silencing of ISG15 has been reported to increase HCV replication [Jones et al., 2010]. Nonetheless, recent reports also suggest that ISG15 promotes HCV production [Chanda et al., 2003; Chua et al., 2009; Broering et al., 2010], suggesting that HCV may exploit ISG15 as part of an immune evasion mechanism [Chen et al., 2010]. ISG15 expression was significantly higher in patients with substitutions at core amino acid 70 and 91, although core substitutions were not significantly associated with outcome of therapy in this study. However, other studies have reported an association between viral substitutions and response to treatment [Enomoto et al., 1996; Akuta et al., 2006b].
Although ISG expression appears to be related to a polymorphism in the IL28B locus, the mechanism has yet to be identified. One proposal is that the favorable genotype disrupts chromatin accessibility or binding of transcription factors, resulting in decreased expression of IL28B, which in turn reduces expression of genes responsive to IL28B [Rauch et al., 2010]. Although this could lower baseline expression levels and help establish a stronger induction during interferon therapy, it does not explain the higher rates of spontaneous clearance associated with the favorable SNP genotype [Thomas et al., 2009]. Alternatively, the effect of the SNP may be more indirect. For example, the favorable SNP genotype is also associated with significantly higher levels of low-density lipoprotein and apolipoprotein B [Li et al., 2010]. In the related West Nile Virus, viral proteins disrupt cholesterol metabolism, which in turn disrupts interferon signaling by down-regulating the Jak-STAT signaling pathway [Mackenzie et al., 2007]. Therefore, it appears that interferon sensitivity may be influenced by a number of viral and host factors. Further research is needed to identify the functional IL28B SNP and elucidate its effect on gene expression and interaction with viral proteins.
The relationship between interferon lambda and ISG expression is also unclear. Honda et al. [2010]. reported a significant positive correlation between IL28B and ISG expression but with slopes differing by IL28B SNP genotype. They found that ISG expression, but not IL28A/B expression, differed significantly by SNP genotype. On the other hand, expression levels of IL28A/B were 10-fold lower than expression levels of representative ISGs, suggesting a lack of statistical power to detect differences [Honda et al., 2010]. Lack of primer sensitivity due to the high sequence similarity between IL28A and IL28B also makes it difficult to detect differences in expression specific to IL28B [Honda et al., 2010].
Another difficulty in interpreting results of ISG expression data is that the role of interferon-lambda in antiviral defense has not been clearly established. Interferons alpha and lambda induce nearly the same set of target ISGs, but interferon alpha is thought to have a stronger effect, whereas the effect of IFN lambda is thought to be more sustained [Marcello et al., 2006]. Therefore, higher baseline expression of interferon lambda might reduce sensitivity to interferon alpha during therapy. Ribavirin may also play a role in potentiating the action of interferon alpha by augmenting the induction of a distinct subset of ISGs, including IRF7 and IRF9 [Thomas et al., 2010]. Regulation of individual ISGs may therefore involve a complex balance between multiple signaling pathways. It is also possible that a discrepancy could exist between mRNA and protein expression levels in terms of quantity or timing that might complicate interpretation of ISG expression data.
In summary, polymorphisms within the IL28 locus appear to be associated with differences in ISG expression levels. The protective IL28B SNP genotype is associated with decreased expression of genes that promote antiviral state and increased expression of genes that suppress antiviral state. Expression levels of antiviral and suppressor ISGs, in turn, appear to be associated independently with the outcome of therapy. Functional analysis of ISGs and IL28 polymorphisms should be performed to clarify the mechanism underlying the differential response to combination therapy for chronic HCV.
Acknowledgements
The authors thank Rie Akiyama for the excellent technical assistance, and Sakura Akamatsu, Mika Tsuzuno, Sanae Furuya, and Aya Furukawa for secretarial assistance. Part of this work was carried out at the Analysis Center of Life Science, Hiroshima University.




