Comparison of the PapilloCheck® assay with the digene HC2 HPV DNA assay for the detection of 13 high-risk human papillomaviruses in cervical and anal scrapes
Abstract
The PapilloCheck® assay was compared with the Digene HC2 HPV DNA assay for the detection of 13 high-risk human papillomaviruses (HPV) in 240 samples, including 181 cervical scrapes and 59 anal scrapes. Overall, 75 (30.5%) samples were positive by the Digene HC2 HPV DNA assay: 34 (18.8%) cervical scrapes and 41 (69.5%) anal scrapes. By considering only the 13 high-risk HPV types detected by the Digene HC2 HPV DNA assay, 66 (27.5%) samples were positive by the PapilloCheck® assay: 27 (14.9%) cervical scrapes and 39 (66.1%) anal scrapes. Concordant results between the two assays were obtained for 225 (93.8%) samples with a Kappa coefficient value of 0.85, indicating an excellent agreement. By considering all the HPV types detectable by the PapilloCheck® assay, the overall prevalence of HPV was 34.2% (82/240): 21.0% (38/181) in cervical scrapes and 74.6% (44/59) in anal scrapes. Among the samples positive by the PapilloCheck® assay, a multiple HPV infection (2–9 HPV types) was identified in 43 of 82 (52.4%) samples, including 7 of 38 (18.4%) cervical samples, and 36 of 44 (81.8%) anal samples. The prevalence of high-risk HPV, as determined by the PapilloCheck® assay, was 17.6% (36/205) in samples with normal cytology, 83.9% (26/31) in samples with low-grade squamous intraepithelial lesions or atypical squamous cells of undetermined significance, and 100% (4/4) in samples with high-grade squamous intraepithelial lesions. The results obtained indicate that the PapilloCheck® assay may be considered as a reliable screening test for HPV detection and typing. J. Med. Virol. 83:1377–1382, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Infection with human papillomaviruses (HPV) is the most common sexually transmitted infection worldwide. Some HPV types, designed as high-risk HPV, have oncogenic properties, and persistent infection with these viruses may lead to the development of precancerous lesions that may evolve into invasive carcinoma [Muñoz et al., 2006]. High-risk HPV are involved in virtually all cases of cervical cancer [Walboomers et al., 1999] and in 25–100% of the other anogenital or head and neck cancers, according to the grade and localization of the lesions [Kreimer et al., 2005; De Vuyst et al., 2009].
Detection of high-risk HPV DNA in cervical scrapes allows to identify women at risk of cervical malignancies and it is now established that HPV DNA testing is a powerful tool in cervical cancer screening [Cuzick et al., 2008; Meijer et al., 2008]. HPV DNA testing in anal scrapes has also potential interest for identifying subjects at risk of anal cancer [Berry et al., 2009; Goldstone et al., 2009].
The Digene HC2 HPV DNA assay (Qiagen, Courtaboeuf, France) is based on a chemiluminescent reaction in which HPV DNA binds to an RNA cocktail probe to detect 13 high-risk HPV types: HPV 16, HPV 18, HPV 31, HPV 33, HPV 35, HPV 39, HPV 45, HPV 51, HPV 52, HPV 56, HPV 58, HPV 59, and HPV 68. This test has shown an excellent sensitivity for the detection of precancerous lesions of the cervix [Arbyn et al., 2006], and the Digene HC2 HPV DNA assay is the only HPV test approved by the US Food and Drug Administration (FDA) for cervical cancer screening. However, this test is not designed for the detection of other high-risk or probably high-risk HPV types such as HPV 26, HPV 53, HPV 66, HPV 73, and HPV 82 [Muñoz et al., 2003, 2006]. Furthermore, the Digene HC2 HPV DNA assay test does not distinguish individual HPV types. It is well established that there is a difference in the carcinogenic potential between the different high-risk HPV types [Wheeler et al., 2006]. In particular, HPV 16 and 18 types have the highest relative risks of cervical cancer and account for 70% of all cervical cancer cases [Muñoz et al., 2004; Khan et al., 2005; Smith et al., 2007; Wheeler et al., 2009; Matsumoto et al., 2010]. A similar situation is observed in HPV anal infection and anal cancer [Hoots et al., 2009]. Therefore, HPV tests that distinguish HPV types may identify individuals at the greatest risk of precancerous or cancerous lesions.
The PapilloCheck® test (Greiner Bio-one, Frickenhausen, Germany) is a PCR-based DNA microarray system that allows to identify 24 HPV types, including the 13 high-risk types detected by the Digene HC2 HPV DNA assay, the high-risk types HPV 73 and HPV 82, the probably high-risk types HPV 53 and HPV 66, and the low-risk types HPV 6, HPV 11, HPV 40, HPV 42, HPV 43, HPV 44/55, and HPV 70 (Fig. 1).
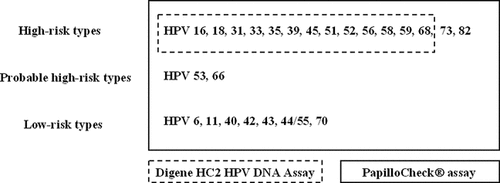
HPV types detected by the Digene HC2 HPV DNA assay and the PapilloCheck® assay.
The aim of the present study was to compare the Digene HC2 HPV DNA and PapilloCheck® assays for the detection of HPV in cervical and anal scrapes.
MATERIALS AND METHODS
Clinical Specimens
Cervical scrapes were collected from pregnant women who benefited from cervical cytology and HPV DNA testing during their pregnancy. Their mean (±SD) age was 26.6 (±4.4) years. Anal scrapes were collected from HIV-1-seropositive men who have sex with men who benefited from anal cancer screening. Their mean (±SD) age was 43.5 (±7.8) years. Cervical scrapes were collected using a Cervex® brush (Cytyc, Montrouge, France) and anal scrapes were collected using a Scrinet® 3 mm brush (Laboratoire CDD, Paris, France). Brushes were agitated vigorously in a vial of PreservCyt transport solution (Cytyc), and the resulting cell suspension was stored at room temperature until analysis.
Cytological Analysis
Thin layer cytological smears were prepared from the cell suspension vial by controlled membrane transfer technology using the ThinPrep 2000 processor (Cytyc). After Papanicolaou staining, slides were examined by a pathologist and interpreted based on the Bethesda 2001 system criteria established for cervical cytology [Smith, 2002]. Cervical and anal smears were interpreted as normal, atypical cells of undetermined significance, low-grade squamous intraepithelial lesions, or high-grade squamous intraepithelial lesions.
Digene HC2 HPV DNA Assay
The Digene HC2 HPV DNA assay was carried out on a 4-ml aliquot of the residual cell suspension according to the manufacturer's instructions. A relative light unit (RLU) cut-off value of 1.0 or greater was considered positive for high-risk HPV detection.
PapilloCheck® Assay
The PapilloCheck® assay is based on the PCR amplification of a DNA fragment of about 350 nucleotides within the E1 gene of HPV. The primers used also generate a PCR product from the PCR control template which is present in the assay mastermix. An internal PCR control is included which targets a region within the human housekeeping gene ADAT1 (adenosine deaminase tRNA specific 1) which is amplified and fluorescence-labeled with Cy5 simultaneously in the same reaction. The amplification products are then hybridized to specific DNA probes fixed on the DNA chip. Each HPV type is detected by a specific DNA probe present in five replicates in each chip. After hybridization and subsequent washing, the chip is scanned with the CheckScanner™ (Greiner Bio-one) at excitation wavelenghts of 532 and 635 nm.
Sample testing using the Papillocheck® assay was carried out following the manufacturer's recommendations. Briefly, DNA was extracted from 100 µl of the residual cell suspension using the MagNa Pure Compact Nucleic Acid Isolation kit (Roche Diagnostics, Meylan, France) on a MagNa Pure Compact automated nucleic acid extractor, with a final elution in 200 µl of elution buffer. PCR was carried out using 5 µl of DNA extract in a final volume of 25 µl with the PapilloCheck® mastermix and 1 U of HotStart Taq DNA polymerase (Qiagen). PCR conditions were: 15 min at 95°C followed by 40 cycles of 30 sec at 95°C, 25 sec at 55°C, and 45 sec at 72°C, followed by 15 cycles of 30 sec at 95°C and 45 sec at 72°C. For hybridization, 5 µl of the PCR product was mixed with 30µl of the hybridization buffer and 25 µl of the mix were transferred into a compartment of the chip. The chips were washed three times with the washing solution at room temperature (10 sec), 50°C (60 sec), and then at room temperature (10 sec). After drying by centrifugation, these chips were scanned using the Checkscanner™. Data were analyzed and results interpreted with the Checkreport™ version 3.0.1 software.
Sequencing
Samples giving a discordant result between the two assays were analyzed by sequencing. A fragment of the L1 gene was amplified by PCR using the PGMY consensus primers [Gravitt et al., 2000]. The amplification mixtures contained 10 pmol of each oligonucleotide in the upstream and downstream primer sets in the presence of ×1 PCR buffer, 4 mM MgCl2, 200 µmol (each) dNTPs, 1.5 U of AmpliTaq gold DNA polymerase (Applied Biosystems, Courtaboeuf, France) and 5µl of DNA extract in a volume of 50 µl. The PCR conditions were 95°C for 9 min followed by 40 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min, and a final extension at 72°C for 5 min. PCR products were analyzed by electrophoresis on a 2% agarose gel stained with ethidium bromide. PCR products of the expected size (450 bp) were sequenced. The GP5+ and GP6+ primers [de Roda Husman et al., 1995] served as forward and reverse sequencing primers, respectively. Sequencing was performed using the BigDye® Terminator v3.1 cycle sequencing kit (Applied Biosystems) on an ABI PRISM® 3500 genetic analyzer (Applied Biosystems). Sequences were aligned using the Lasergene® sequence analysis software (DNAstar, Madison, WI) and then compared through BLAST to HPV reference sequences using GenBank database (http://www.ncbi.nlm.nih.gov/blast). HPV type identification was based on a sequence similarity with an identified HPV type >90%.
Data Analysis
When the Digene HC2 HPV DNA assay was positive, the two assays were considered as concordant if one or more of the 13 HR-HPV types detected by the Digene HC2 HPV DNA assay was identified by the PapilloCheck® assay. When the Digene HC2 HPV DNA assay was negative the two assays were considered as concordant if none of these 13 HR-HPV types was identified by the PapilloCheck® assay.
Agreement between the two tests was assessed by the Cohen's kappa coefficient, with values of 0.00–0.20 indicating poor agreement, 0.21–0.40 indicating fair agreement, 0.41–0.60 indicating moderate agreement, 0.61–0.80 indicating good agreement, and 0.81–1.00 indicating excellent agreement.
The prevalences of high-risk HPV DNA detection, multiple HPV infection and cytological abnormalities were compared between cervical and anal specimens using the χ2 test. P < 0.05 was considered as significant.
RESULTS
A total of 240 samples, including 181 cervical scrapes and 59 anal scrapes were analyzed by the two assays. Overall, 75 (30.5%) samples were positive with the Digene HC2 HPV DNA assay: 34 (18.8%) cervical scrapes and 41 (69.5%) anal scrapes. By considering only the 13 high-risk HPV types detected by the Digene HC2 HPV DNA assay, 66 (27.5%) samples were positive by the PapilloCheck® assay: 27 (14.9%) cervical scrapes and 39 (66.1%) anal scrapes. By considering all the HPV types detectable by the PapilloCheck® assay, the overall prevalence of HPV was 34.2% (82/240): 21.0% (38/181) for cervical scrapes and 74.6% (44/59) for anal scrapes. The HPV type distribution is presented in Figure 2. Among the samples positive by the PapilloCheck® assay, a multiple HPV infection (2–9 HPV types) was identified in 43 of 82 (52.4%) samples: 7 of 38 (18.4%) cervical scrapes and 36 of 44 (81.8%) anal scrapes.
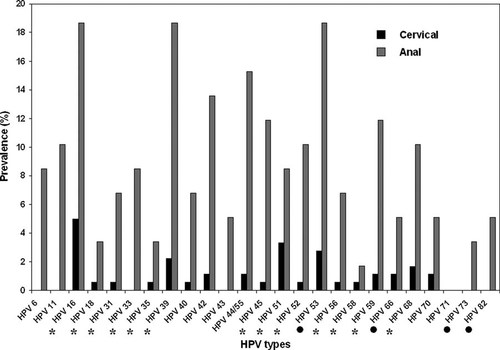
Distribution of HPV types in cervical (n = 181) and anal (n = 59) scrapes as detected by the PapilloCheck® assay. Asterisks indicate the 13 high-risk HPV types targeted by the Digene HC2 HPV DNA assay. Solid circles indicate additional high-risk or probably high-risk HPV types.
As shown in Table I, 225 (93.8%) samples gave concordant results in both assays, with a Kappa coefficient value of 0.85, indicating an excellent agreement. Samples giving a discordant results are presented in Table II. Genotype identification determined by sequencing in the discordant samples was in agreement with the results obtained by the PapilloCheck® assay.
| PapilloCheck | ||
|---|---|---|
| Positive | Negative | |
| HC2 | ||
| Positive | 63 | 12 |
| Negative | 3 | 162 |
| Sample no. | HC2 | PapilloCheck | Sequencing | Cytology |
|---|---|---|---|---|
| Cervical | ||||
| C27 | Positive | HPV 53 | ND | Normal |
| C38 | Positive | Negative | Negative | Normal |
| C45 | Positive | HPV 53 | HPV 53 | Normal |
| C55 | Positive | HPV 66 | HPV 66 | LSIL |
| C82 | Positive | HPV 53 | HPV 53 | Normal |
| C105 | Negative | HPV 58 | ND | Normal |
| C136 | Positive | HPV 53, HPV 70 | HPV 53 | LSIL |
| C141 | Negative | HPV 16 | HPV 16 | Normal |
| C145 | Positive | Negative | HPV 66 | Normal |
| C162 | Positive | Negative | ND | Normal |
| C165 | Positive | HPV 42 | HPV 42 | Normal |
| Anal | ||||
| A13 | Negative | HPV 31, HPV 73 | ND | Normal |
| A29 | Positive | HPV 53, HPV 66 | HPV 66 | ASCUS |
| A39 | Positive | HPV 53 | HPV 53 | Normal |
| A50 | Positive | HPV 66 | HPV 66 | Normal |
- ASCUS, atypical squamous cells of undetermined significance; LSIL, low-grade squamous intraepithelial lesions; ND, not done.
Among the cervical scrapes, cytological abnormalities were observed in 8 (4.4%) samples: 3 (1.7%) atypical squamous cells of undetermined significance, 4 (2.2%) low-grade squamous intraepithelial lesions, and 1 (0.5%) high-grade squamous intraepithelial lesions. Among the anal scrapes, cytological abnormalities were observed in 27 (45.8%) samples: 6 (10.2%) atypical squamous cells of undetermined significance, 18 (30.5%) low-grade squamous intraepithelial lesions, and 3 (5.1%) high-grade squamous intraepithelial lesions. The prevalence of high-risk HPV (13 types), as determined by the PapilloCheck® assay, was 17.6% (36/205) in samples with normal cytology, 83.9% (26/31) in samples with low-grade squamous intraepithelial lesions or atypical squamous cells of undetermined significance, and 100% (4/4) in samples with high-grade squamous intraepithelial lesions. As shown in Table III, the prevalence of high-risk HPV in samples with normal cytology was much higher in anal scrapes than in cervical scrapes (P < 0.0001). Cytological abnormalities were observed in 3 of 12 (25%) samples found positive by the Digene HC2 HPV DNA assay but negative by the PapilloCheck® assay for the detection of the HPV types targeted by the Digene HC2 HPV DNA assay (Table II).
| Cytology | Samples | |
|---|---|---|
| Cervical | Anal | |
| Normal | 21/173 (12.1%) | 15/32 (46.9%) |
| LSIL/ASCUS | 5/7 (71.4%) | 21/24 (87.5%) |
| HSIL | 1/1 (100%) | 3/3 (100%) |
- ASCUS, atypical squamous cells of undetermined significance; LSIL, low-grade squamous intraepithelial lesions; HSIL, high-grade squamous intraepithelial lesions.
The differences observed between cervical and anal scrapes in the prevalence of high-risk HPV DNA detection, multiple HPV infection, and cytological abnormalities were highly significant (P < 0.0001 for all three variables).
DISCUSSION
In the present study, the PapilloCheck® assay was compared with the FDA-approved Digene HC2 HPV DNA assay for the detection of 13 high-risk HPV types in cervical and anal scrapes. An excellent agreement (93.8%) between the two assays was observed. This is in accordance with other studies that reported a high agreement between the PapilloCheck® assay and the Digene HC2 HPV DNA assay [Dalstein et al., 2009; Halfon et al., 2010; Schopp et al., 2010] or other HPV tests such as the linear array [Dalstein et al., 2009; Halfon et al., 2010], the GP5+/6+ PCR enzyme immunoassay [Jones et al., 2009; Hesselink et al., 2010], the PGMY09/11 PCR line blot assay and the SPF10 PCR line probe assay [Schopp et al., 2010]. The discrepant results were due mainly to a cross-reactivity of the Digene HC2 HPV DNA assay with HPV types other than those theoretically detected by the assay. Indeed, the Digene HC2 HPV DNA assay was positive in samples in which HPV 53, HPV 66, or HPV 42 were detected by both the PapilloCheck® assay and sequencing. A cross-reactivity of the Digene HC2 HPV DNA assay with these HPV types as well as with other oncogenic or non-oncogenic HPV types such as HPV types 26, 61, 67, 71, or 81 has been previously reported [Castle et al., 2002, 2008; Poljak et al., 2002]. Three other cervical samples were positive by the Digene HC2 HPV DNA assay but negative by the PapilloCheck® assay, one of them being positive for HPV 66 by sequencing, another one being negative by sequencing, whereas the third sample could not be tested by sequencing.
In the present study, cross-reactivity in the Digene HC2 HPV DNA assay was observed mainly with HPV 53 and HPV 66. Given the fact that these two HPV types are considered now as probably oncogenic [Muñoz et al., 2006], this cross-reactivity is not a real problem for cervical or anal cancer screening. Indeed in the present study, cytological abnormalities were observed in three samples giving a cross-reactivity with HPV 53 and/or HPV 66.
As expected, results obtained from cervical and anal samples were highly different. Indeed, the prevalences of high-risk HPV detection, multiple HPV infection, and cytological abnormalities were considerably higher in the anal scrapes than in the cervical ones. This difference reflects the fact that HIV-1-infected men who have sex with men are at high risk of anal HPV infection and HPV-associated anal precancerous lesions [Chin-Hong et al., 2008; Damay et al., 2010]. The high prevalence of high-risk HPV in anal samples with normal cytology observed in the present study is in agreement with previous studies [Fox et al., 2005].
In summary, results from the present study demonstrate a high agreement between the Digene HC2 HPV DNA assay and the PapilloCheck® assay for the detection of 13 high-risk HPVs. However, cross-reactivities with untargeted HPV genotypes may be observed with the Digene HC2 HPV DNA assay. The main advantages of the PapilloCheck® assay is HPV type identification, detection of more HPV oncogenic and non-oncogenic types as compared with the Digene HC2 HPV DNA assay, as well as identification of multiple HPV infections. The risk of progression of HPV-associated lesions to precancerous or cancerous lesions varies with HPV types [Khan et al., 2005; Wheeler et al., 2009; Matsumoto et al., 2010]. Therefore, HPV genotyping is useful for identifying patients at increased risk of cancer development [Meijer et al., 2006]. The PapilloCheck® assay may thus be considered as a reliable screening test for HPV detection and typing.




