Monitoring of group C rotavirus in children with acute gastroenteritis in Brazil: An emergent epidemiological issue after rotavirus vaccine?†
Conflict of interest: none declared.
Abstract
Group C rotavirus (GpCRV) has a worldwide distribution; however, its epidemiology and ecology are still unclear. Evidence for a possible zoonotic role has been postulated recently for Brazilian children strains. The aim of this study was to monitor GpCRV in children ≤15 years with acute gastroenteritis during the 2007–2010 national Brazilian rotavirus surveillance, and to undertake the molecular characterization of the major VP6 capsid protein. A total of 3,019 fecal samples were first screened for Group A rotavirus (GpARV). A total of 2,205 GpARV ELISA negative samples were tested further for the presence of GpCRV by SDS–PAGE, electronic microscopy, and RT-PCR for the VP6 gene. The genetic diversity of GpCRV was carried out by sequencing the VP6 gene. GpARV and GpCRV infections were detected in 24.6% (742/3,019) and 0.3% (8/3,019), respectively. The GpCRV detection rate increased from 0.2% (1/422) in 2007 to 1% (7/708) in 2008, and GpCRV cases were not detected in 2009 and 2010. The phylogenetic analysis indicated that the strains belonged to the human lineage, and showed a genetic relationship with the GpCRV strain from Japan isolated in 2009. None of the study sequences was related closely to animal GpCRV strains. This study provides further evidence that GpCRV is a minor cause of acute childhood gastroenteritis in Brazil, and does not suggest that GpCRV may assume epidemiological importance in the future, even after the introduction of a GpARV vaccine. In addition, the molecular analyses of the GpCRV samples in this study do not support the zoonotic hypothesis. J. Med. Virol. 83:1631–1636, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
The Group C rotavirus (GpCRV) was first detected from piglets in 1980 [Saif et al., 1980] and confirmed later as a human pathogen by Bridger et al. [1986]. Since then, GpCRV have been detected from both sporadic episodes and outbreaks of gastroenteritis throughout the world [Cunliffe et al., 2001; Phan et al., 2004; Schnagl et al., 2004; Steyer et al., 2006; Abid et al., 2007]. The GpCRV infection in Brazil was first detected in the city of Rio de Janeiro by Pereira et al. [1983]. Thereafter, the GpCRV infections have been identified sporadically or as outbreaks at various times and locations [Gabbay et al., 1989, 1999; Timenetsky et al., 1993; Souza et al., 1998; Teixeira et al., 1998].
There are several unresolved questions about the ecology of the GpCRV. Its epidemiology has not been described fully due to the lack of samples, sensitive diagnostic assays [Castello et al., 2000], and the sporadic pattern of detection. The description of localized outbreaks may be interpreted as a spillover introduction of GpCRV strains from an unidentified source or reservoir [Bányai et al., 2006; Luchs et al., 2009]. A possible zoonotic role has been postulated based on the elevated GpCRV seroprevalence rates in human populations living in rural settings [Iturriza-Gómara et al., 2004]. In fact, Gabbay et al. [2008] documented the carriage of porcine GpCRV in children in Belém, Brazil, and established this virus as another emerging zoonotic infection in humans. The possibility of zoonotic spread of GpCRV has also been considered based on the results of a seroepidemiological survey conducted in the United Kingdom in which the prevalence of antibodies was higher in rural population than in urban population [Iturriza-Gómara et al., 2004].
The GpCRV was detected both in sporadic cases and outbreaks affecting adults and children from rural areas in the states of São Paulo (SP) and Goiás (GO) during the 2007–2008 national Brazilian rotavirus surveillance [Luchs et al., 2009]. This was the first report of GpCRV detection in Southeastern Brazil in over 15-year period of monitoring, however, no genetic information was described in the report. It seems important to improve the GpCRV laboratory surveillance system, especially after 2006 when the RIX4414 rotavirus vaccine was included in the Brazilian Immunization Program, preventing severe rotavirus gastroenteritis, and inducing significant reduction in the frequency of Group A rotavirus (GpARV) detection in children with gastroenteritis [Gurgel et al., 2008]. In the future, the GpCRV may assume an epidemiological importance.
The aim of this study was to intensify the monitoring of GpCRV detection in fecal samples from children ≤15 years with acute gastroenteritis during the 2007–2010 national rotavirus surveillance. In addition undertake the molecular characterization of the major capsid protein (VP6) of the GpCRV strains detected from children during this period, in order to obtain further information on the genetic relationships between human and animal.
MATERIALS AND METHODS
Samples
The aim of the Acute Diarrhea Disease Monitoring Program (ADDM), with a national range is the early detection of diarrhea outbreaks. Stool samples from patients with acute gastroenteritis are sent to Enteric Diseases Laboratory of the Adolfo Lutz Institute, reference center for rotavirus surveillance and a member of ADDM, in order to conduct the viral investigation. The clinical samples tested in this study were obtained from the following states: São Paulo (SP), Mato Grosso do Sul (MS), Paraná (PR), Tocantins (TO), Goiás (GO), Santa Catarina (SC), and the Brazilian Federal District (FD) (Fig. 1). The samples studied were part of ADDM, acquired from a convenient retrospective sampling, without inclusion or exclusion criteria, with no characterization of the participating health centers, and without patients' clinical evaluation. Therefore, the study may not be representative of the actual epidemiological scenario. This retrospective study was conducted from September 2007 to September 2010 with convenient surveillance specimens collected from children under the age of 15 years presenting with acute gastroenteritis.
The states highlighted in black collected stool samples from patients with acute gastroenteritis and sent to the Enteric Diseases Laboratory of the Adolfo Lutz Institute, regional reference center for rotavirus surveillance, member of Acute Diarrhea Disease Monitoring Program and its national range.
Detection of Rotaviruses
A total of 3,019 fecal samples were screened first by a commercial ELISA assay (Premier™ Rotaclone®, Meridian Bioscience, Inc., Cincinnati, OH) performed according to the manufacturer's instructions, for the GpARV detection, the most common rotavirus affecting humans. A total of 2,205 GpARV ELISA negative samples were tested further for the presence of GpCRV by polyacrylamide gel electrophoresis (SDS–PAGE) according to standard procedures, and confirmed by electronic microscopy (EM).
RNA Extraction and RT-PCR
The GpCRV dsRNA was extracted by QIAamp® Viral RNA Mini kit (Qiagen, Inc., Valencia, CA) according to the manufacturer's instructions, and it was stored at −20°C. RT-PCR protocol and the set of primers (C1, C3, and C4) used to amplify a fragment of the VP6 gene was described by Gouvea et al. [1991]. The expected length for RT-PCR products, synthesized with C1–C4 (nt 994–1349), and C1–C3 (nt 994–1320) pairs were 356 and 327 bp, respectively.
Sequence and Phylogenetic Analysis
The RT-PCR products were purified with PureLink™ Purification kit (Invitrogen, Carlsbad, CA). Cycle sequencing was carried out using the BigDye kit v1.1 (Applied Biosystems, Foster City, CA) with primers C1, C3, and C4 specific for the VP6 gene. Dye-labeled products were run on an automated sequence analyzer (ABI Prism type 3100; Applied Biosystems). All sequencing chromatograms obtained were edited manually to obtain contigs, using Sequencher 4.7 software. All sequences were screened at the NCBI website using the Basic Local Alignment Search Tool (BLAST). Sequences generated by manual edition and a set of cognate sequences of human and animal GpCRV, available in the GenBank database, were aligned using Clustal W program [Thompson et al., 1994]. Minor manual adjustments were made to improve the alignment using BioEdit software (Ibis Therapeutics, Carlsbad, CA). Phylogenetic analysis was performed with PAUP4b10 [Swofford et al., 1996]. Neighbor Joining (NJ) and Maximum Likelihood (ML) trees were constructed based on K80 substitution model with a gamma distribution shape parameter, determined by Modeltest v3.7 [Posada and Crandall, 1998]. Bootstrap was assessed using 1,000 replicates. Trees were visualized using Tree View program [Page, 2002].
The genetic distance of the VP6 amino acid sequences were estimated for all the sequences included in this study. Mean genetic distances within and between groups were determined using Mega v4.0 program [Kumar et al., 2001]. Genetic distances were estimated with the Poisson correction model.
Nucleotide Sequences Accession Numbers
The nucleotide sequences determined in this study have been deposited in the GenBank database and assigned the accession numbers HM853819–HM853824.
RESULTS
Detection of Rotaviruses
The GpARV infection was detected in 24.6% (742/3,019) of the samples, being 26% (110/422) in 2007, 30.2% (214/708) in 2008, 2.8% (19/681) in 2009, and 33% (399/1,207) in 2010. The median age for patients infected with the GpARV was 2.4 years old. The GpCRV infection was detected in eight samples: 0.4% of the subset tested (8/2,205) and 0.3% of the overall study (8/3,019). The GpCRV samples were detected in five gastroenteritis outbreaks and three sporadic cases; the median age was 4.5 years old; 75% males (Table I). Concomitants GpARV infections, occurring in some outbreaks, were also observed, however, no co-infection was detected. The GpCRV detection rate increased from 0.2% (1/422) in 2007 to 1% (7/708) in 2008, and GpCRV cases were not detected in 2009 and 2010.
| Age (years) | Sex | Municipality | State | Month/year | Case | Strains | Accession numbers |
|---|---|---|---|---|---|---|---|
| 2 | M | Ipeúna | SP | December 2007 | Sporadic | SP22/Hu/BR/2007 | HM853819 |
| 2 | M | Catanduva | SP | March 208 | Outbreak | SP09/Hu/BR/2008 | HM853820 |
| 7 | M | Catanduva | SP | March 2008 | Outbreak | SP12/Hu/BR/2008 | HM853821 |
| 7 | M | Sales | SP | May 2008 | Outbreak | SP28/Hu/BR/2008 | HM853822 |
| 6 | M | Salto | SP | July 2008 | Outbreak | –– | |
| 10 | F | Pontalinda | SP | August 2008 | Sporadic | SP35/Hu/BR/2008 | HM853823 |
| 1 | F | Mauá | SP | September 2008 | Sporadic | SP36/Hu/BR/2008 | HM853824 |
| 1 | M | Pirineus | GO | September 2008 | Outbreak | –– |
The seven samples from SP and the one sample from GO showed a similar electrophoretic profile, and the characteristic 4-3-2-2 genomic migration pattern compared with the SA-11 strain of the GpARV dsRNA (data not shown). Electron microscopy analysis revealed both complete rotavirus and damage rotavirus-like particles in samples containing GpCRV.
RT-PCR Amplification and Phylogenetic Analyses of the VP6 Genes
Of the eight GpCRV positive samples, six specimens were amplified successfully by the RT-PCR assay, and their sequences were determined. All the six GpCRV strains (SP22/Hu/BR/2007, SP09/Hu/BR/2008, SP12/Hu/BR/2008, SP28/Hu/BR/2008, SP35/Hu/BR/2008, SP36/Hu/BR/2008) belong to SP and are listed in Table I.
All the GpCRV SP strains were very similar to the GpCRV samples on BLAST search based on the VP6 protein. The phylogenetic analysis indicated that the GpCRV SP strains belonged to the human lineage. None of the GpCRV SP sequences were related closely to animal GpCRV strains (Fig. 2). The GpCRV SP strains were related to each other, since they shared 97–100% amino acid (aa) identities in the VP6 gene.
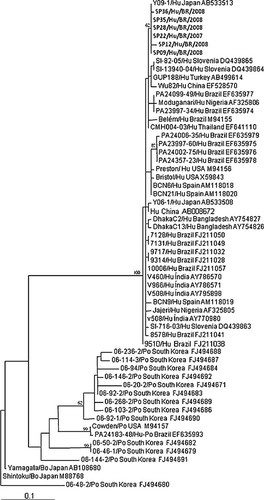
Maximum Likelihood phylogenetic tree of the partial VP6 nucleotide sequences generate with K80 substitution model plus gamma distribution shape parameter of the six human GpCRV strains isolated in the state of São Paulo. References of the GpCRV strains were obtained from GenBank database. Percentage of bootstrap values greater than 60 are shown at the branch node. The strains isolated from the state of São Paulo State are indicated in boldface. The location and accession number of each strain are indicated.
The GpCRV SP strains analyzed in this study showed a genetic relationship with GpCRV strains from Japan, especially from the city of Yokohama (AB533513) isolated in 2009. The other humans GpCRV Brazilian strains available in GenBank clustered with strains from Nigeria (AF325806), Thailand (EF641110), United Kingdom (M94156, X59843), Spain (AM118018, AM118020), India (AY786570, AY786571, AY795898), and Slovenia (DQ439863). In addition, 12 porcine Korean GpCRV strains were placed on separated branches from the human GpCRV strains, Cowden strain and porcine-like human Brazilian children strains (Fig. 2).
Genetic Distance Analysis
The VP6 amino acid sequences were grouped in bovine GpCRV, porcine GpCRV, human GpCRV, and GpCRV SP samples for the distance analysis. The intragroup distance was 3.2%, for bovine group, 21.9% for porcine, 7% for human, and 1.1% for the study samples. Once observed higher distance in the porcine and human groups, another analysis was conducted excluding the prototype porcine Cowden sequence of the porcine group and separating the human sequences in different groups: a group of human sequences from Brazil, a group of human sequences from Japan, and another group for the remaining human sequences.
This newer analysis showed a genetic distance of 21.1% for porcine group, 5% for Brazilian sequences, 6.6% for Japanese, and 4.5% for the remaining human sequences. The higher value of genetic distance obtained in the first analysis for human sequences were possibly due to the presence of the sequences isolated in children from the city of Belém with a possibly porcine origin.
The genetic distance between these latest groups showed that the study sequences have the same genetic distance for bovine (50.1%), porcine Cowden (54.1%), porcine (54.5%), and human porcine-like Belém samples (54.1%). The genetic distance among Japanese samples were 3.8%.
The genetic distance between bovine and porcine group were 29.4%. Among the bovine and the porcine Cowden sequence were 26.2%, similar to the observed between porcine group and porcine Cowden sequence, 26.3%.
DISCUSSION
This study was designed in order to investigate the frequency of GpCRV infection in children ≤15 years old with acute gastroenteritis and addressed the genetic relationship of the GpCRV detected with others human, porcine, and bovine strains. The frequency of GpCRV infections detected in this work (0.3%) was similar to that observed in other studies carried out with pediatric population in South Korea (0.7%) [Moon et al., 2011] and Turkey (0.8%) [Mitui et al., 2009]. However, the prevalence of the infection was lower compared to Nigeria (1.8%) [Adah et al., 2002], Argentina (2.8%) [Castello et al., 2002], Spain (16%) [Sánchez-Fauquier et al., 2003], Malawi (3.3%) [Cunliffe et al., 2001], and Japan (1.2%) [Kuzuya et al., 2007]. This study does not suggest that the GpCRV have currently a major epidemiological impact, even after the introduction of a GpARV vaccine. Indeed, the decrease in the detection has fallen to zero in 2009 and 2010.
Considering the GpARV detection, the annual frequency was similar to that reported in studies carried out in Brazil [Linhares, 2000; Carmona et al., 2006]; however, a low percentage of GpARV infection was observed in 2009. The Brazilian States of Midwest, Southeast, and South regions exhibit a temperate-like climate, with positive GpARV specimens appearing to peak during the winter or dry season (from May to September) [Pereira et al., 1993]. According to the National Meteorology Institute (INMET) records, during the year of 2009, Brazil experienced an atypical raining winter season with higher average rainfall record due to the El Niño phenomenon (http://www.inmet.gov.br) therefore, this climatic condition could be involved in the low detection frequency observed.
The median age of GpARV and GpCRV detections in the current study were 2.4 and 4.5 years old, respectively, agreeing with seroepidemiological investigations and detection data that suggest a difference between age distribution in children with diarrhea caused by GpARV, which typically infects children before 3 years of age, and GpCRV, which affects older children [Nilsson et al., 2000; Kuzuya et al., 2001; Iturriza-Gómara et al., 2004; Castello et al., 2009].
Seven of eight GpCRV samples were detected in SP, suggesting that the GpCRV occurs more frequently in this state. However, this may also be an outcome from the efficient health surveillance program conducted in the state, and the higher number of samples sent to laboratory for viral diagnosis. Serological investigations also suggest a higher GpCRV incidence in rural than in urban populations [Iturriza-Gómara et al., 2004; Mukhopadhya et al., 2010]. A similar pattern was observed in this study, once the eight samples were collected from people who live in countryside areas. However, there is no serological information about GpCRV from diverse geographical areas in Brazil, and the common source of these infections was unable to be identified during the study.
The high aa identity level of the GpCRV SP strains in the VP6 protein, suggest that these strains were originated from a single or a group of related strains. Also, the partial VP6 gene analyzed showed a relationship between the GpCRV strains from SP and Japan, especially from the city of Yokohama, Kanagawa prefecture. Currently, Brazil is the country with the largest number of Japanese outside Japan, and the Brazilian community in Japan for its part, is the third largest community of foreign workers residing in Japan [Beltrão and Sugahara, 2006]. The fact that this migration flow may lead to a possible exchange of GpCRV strains can be speculated, however, additional analysis of full-length of the VP6 gene, and/or VP7 and VP4 genes are required to fully validate this finding.
The molecular analysis showed that human strains were related distantly to the bovine and porcine strains as observed by Mitui et al. [2009]. The human GpCRV strains appear to constitute a single linage supported by high bootstrap values, irrespective of the geographic origin. The highly conservation among the human GpCRV genetic sequences could suggest a recent evolution.
In this study, all samples from SP cluster with other human samples and did not show any evidence of animal ancestry. However, the human porcine-like samples from Belém clustered with the prototype porcine Cowden in agreement with the data showed by Gabbay et al. [2008]. There are evidences that the transmission of the GpARV can occur from animal-to-human as well as from animal-to-animal by direct transmission of the virus or the contribution of one or several genes to reassortants [Martella et al., 2006; Ghosh et al., 2007]. Compared to the GpARV, there is a paucity of information regarding the sequence and phylogenetic data on all 11 genomic segments of the GpCRV. Therefore, it is largely unknown whether these genomic segments are totally similar to those of the original species or reassortants from other species. However, the GpCRV are thought to be able to cause interspecies transmission [Jeong et al., 2009].
In fact, the oldest GpCRV samples were collected from pigs, which strongly suggest that the origin of this pathogen could be porcine. The acquisition of genes with human origin could facilitate the adaptation of such strain in order to infect the human population. The fact that the GpCRV strains do not remain circulating in humans (like GpARV), may suggest that this group does not achieve the fitness required to become a successful human pathogen. An alternative hypothesis is that the GpCRV may cause a subclinical infection in humans. Based on Meleg et al. [2008] study conducted in Hungary, the GpCRV were detected from raw sewage samples, suggesting that the virus is in circulation; however, a significant increase in the number of sporadic cases or outbreaks was not observed.
In conclusion, this study adds further evidence that the GpCRV is a minor cause of acute childhood gastroenteritis in Brazil during the surveillance period. In addition, small-scale of GpCRV outbreaks in restricted communities might occur and go undetected, particularly if the severity of the disease is minor and hospitalization is not required [Bányai et al., 2006]. It is noted that the GpCRV incidence and associated disease remain unclear once sensitive tests for its detection are not available to clinical laboratories. The diagnosis is difficult since most of the ELISA assays do not recognize the GpCRV specific VP6 antigen. The RT-PCR using GpCRV specific primer is a convenient option [Castello et al., 2000]; however, it has not been used widely due to the large costs for routine surveillance. The PAGE analysis of dsRNA requires the presence of 108–1010 viral particles/ml for a positive result [Castello et al., 2000] nevertheless; this method has sufficient sensitivity to detect GpCRV with low costs.
Acknowledgements
We thank João Leandro de Paula Ferreira and João Paulo Gervasio Batista, Retrovirus Laboratory of Adolfo Lutz Institute for their assistance with the sequencing reaction and edition. We thank Marli Ueda and Jonas José Kisielius, Electronic Microscopy Laboratory of Adolfo Lutz Institute for their assistance with the electronic microscopy analysis. We are especially thankful to Enteric Diseases Laboratory staff, Audrey Cilli for critical comments on the manuscript, Cibele Daniel Ribeiro for technical assistance, Samira Julien Calux for assistance with sample collection, and Rita de Cássia Compagnoli Carmona for laboratory supervision. We thank Camila Buoro Auler for the specialized English language review.




