Greater diversity of HIV DNA variants in the rectum compared to variants in the blood in patients without HAART
Abstract
The gut-associated lymphoid tissue represents the largest reservoir of HIV-1. Improving knowledge of this reservoir by studying the diversity of viral population is a key step towards understanding the pathogenesis and dynamics of HIV. Obtaining samples is difficult and little information is available on gut viral quasispecies during the course of infection in humans. The aim of this study was to characterize rectal viral strains and their diversity and to investigate the relationships between the rectal tissue reservoir and viral variants in the blood. Phylogenetic analyses were performed on the env sequences for rectal HIV DNA, blood HIV DNA, and HIV RNA clones, with maximum-likelihood and neighbor-joining methods on seven patients. Genetic diversity was assessed. Higher diversity of HIV DNA clones was noted in the rectum compared to blood in four out of five patients without HAART. Viral diversity was present in the rectum from time of the primary infection. Similar degrees of diversity were observed in the rectum and blood during HAART. Rectal and blood HIV variants were interspersed partially or totally in the seven patients. A certain number of rectal HIV DNA clones were clustered together in six patients. These results suggest that variants in the rectum were more heterogeneous than variants in the blood from patients without HAART, probably because the activated milieu of gut-associated lymphoid tissue may provide an improved environment for viral replication, and indicate exchange of viral populations between blood and rectal tissues, reflecting the dynamics of HIV during course of infection. J. Med. Virol. 83:1499–1507, 2011. © 2011 Wiley-Liss, Inc.
Abbreviations:
HAART, highly active antiretroviral treatment; HIV, human immunodeficiency virus; PBMCs, peripheral blood mononuclear cells.
INTRODUCTION
The mucosal-associated lymphoid tissue contains 40–60% of the body's lymphocytes [Cerf-Bensussan and Guy-Grand, 1991; Mowat and Viney, 1997]. The gut-associated lymphoid tissue represents the largest reservoir of human immunodeficiency virus type 1 (HIV-1) [Clayton et al., 1997; Veazey et al., 1998; Brenchley et al., 2004; Veazey and Lackner, 2005; Chase et al., 2006]. Although HIV-1 replication decreases in gut-associated lymphoid tissue with highly active antiretroviral treatment (HAART), HIV RNA and replication-competent HIV-1 can be detected in gut-associated lymphoid tissue of patients under successful HAART, thus confirming that the intestinal mucosa can act as a residual HIV reservoir despite HAART [Poles et al., 2006; Belmonte et al., 2007].
Improving knowledge of this intestinal reservoir is a key step towards the understanding of the pathogenesis of HIV and better assessment of the efficacy of treatment. Studying the diversity of the viral population within-, and analyzing the potential differences between blood and the intestine may provide an insight into the dynamics of HIV-1.
Previous studies on HIV quasispecies have described HIV compartmentalization in various tissues and fluids, including the brain, lung, semen, and cervicovaginal secretions [Korber et al., 1994; Zhu et al., 1996; Wong et al., 1997; Delwart et al., 1998; van't Wout et al., 1998; Chomont et al., 2007]. Due to the difficulty of obtaining samples, little information is available on gut viral quasispecies during the course of infection in humans. Colon-derived HIV-1 variants were studied from autopsied samples from a patient who died of AIDS, but these variants were not compared to circulating blood variants [Wang et al., 2001]. Based on Nef analysis, Van Marle et al. [2007] showed compartmentalization of the gut HIV reservoir in patients infected chronically prior to HAART. However, results were less clear with reverse transcriptase sequences. Poles et al. [2001] have observed the presence of some HIV RNA variants resistant to non-nucleosidic reverse transcriptase inhibitor in gut-associated lymphoid tissue and in peripheral blood mononuclear cells (PBMCs) which were different from those observed in the plasma. Evidence of cross-infection between gut-associated lymphoid tissue and PBMCs during HAART has been suggested as a possible mechanism for persistence of HIV [Chun et al., 2008].
In this context, the objective of the study was to characterize rectal viral populations, to compare their diversity to the diversity of blood variants, in patients receiving treatment or not, and to investigate the relationship between viral variants in the rectum and those circulating in PBMCs and in the plasma. This study was based on rectal biopsies because they are the easiest gut biopsies to obtain in humans.
STUDY DESIGN
Patients and Samples
Seven patients, enrolled in a study described previously [Avettand-Fenoel et al., 2008], were examined. Two of the patients were at the time of the primary infection within 4 months of the onset of the acute retroviral illness, four patients were infected chronically and one patient had AIDS (Table I). Blood samples and four punches of rectum including the lamina propria were collected on the same day. In two patients, a second set of samples was taken several months later (B, C) (Table I). In total, six sets of samples were collected in patients without HAART (A, B first set of samples, C first set of samples, C second set of samples, D, G) and three during HAART (B second set of samples, E, F) (Table I). Patients A, B (1st sample), C were naïve to HAART; Patients D and G had previously received HAART but were not receiving treatment at the time of sample collection (Table I). Rectal endoscopic biopsies were undertaken at the same site to avoid potential variations among patients. Rectal tissue was also collected from the same site for histopathological assessment. The local ethics committee (Tours, France) approved the study and patients gave their written informed consent.
| Pt | Sex | Stage of infection | Duration of infection (months) | Zenith HIV RNA Log 10 copies/ml | CD4 T cell nadir cells/mm3 | Treatment (tritherapy) | CD4+ cell count | Plasma HIV RNA, log cp/ml | HIV DNA, Log cp/million cells | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Blood/mm3 | Rectum/million cells | PBMCs | Rectum | ||||||||
| A | M | PI | 3a | 5.8 | 267 | Naïve | 322 | NA | 5.8 | 3.3 | 3.3 |
| B | M | PI | 1a | 5.2 | 729 | Naïve | 729 | NA | 5.1 | 3.6 | 4.0 |
| 3.5a | 5.2 | 729 | For 2.5 months | 850 | NA | <1.7 | 3.2 | 3.9 | |||
| C | M | CI | 22 | 5.7 | 261 | Naïve | 469 | 20,770 | 4.6 | 3.3 | 3.0 |
| 37 | 5.7 | 251 | Naïve | 251 | NA | 5.7 | 3.6 | 3.5 | |||
| D | M | CI | 117 | 4.9 | 377 | No | 425 | 7,310 | 4.3 | 3.7 | 4.0 |
| E | M | CI | 122 | 4.4 | 105 | For 4 years | 270 | NA | <1.7 | 3.5 | 2.8 |
| F | F | CI | 64 | 5.3 | 222 | For 3 years | 426 | 32,890 | <1.7 | 3.5 | 2.8 |
| G | F | AIDS | 120 | 6.0 | 15 | No | 15 | 5,600 | 6.0 | 3.8 | 3.3 |
- M, male; F, female; PI, primary infection; CI, chronic infection; PBMCs, peripheral blood mononuclear cells; cp, copy; no, no treatment; NA, not available.
- a After onset of acute retroviral illness.
Nucleic Acids Extraction
RNA was extracted from plasma using the QIAamp RNA Mini Kit (Qiagen, Courtaboeuf, France) and used to synthesize cDNA with the Reverse transcriptase core kit (Eurogentec, Angers, France). Total DNA was extracted from PBMCs using the QIAamp DNA mini kit and from rectal tissues using the QIAamp tissue kit (Qiagen). The extracts of the four rectal biopsies collected on the same day from each patient were pooled.
Cloning and Sequencing
The env regions encoding C2-V5 were amplified using a previously described nested PCR [Delwart et al., 1993]. Viral sequences were amplified using an Expand High FidelityPLUS PCR system (Roche Diagnostics, Meylan, France). To avoid a PCR-based bias, at least two PCRs from the pooled extracts were performed on each sample. Rectal HIV DNA, blood HIV DNA, and blood HIV RNA from each patient, as well as the sequential samples, were amplified on different days to avoid contamination. To prevent any inter-sample contamination, water and one sample from each patient were amplified on one day. Moreover, the different amplifications of the samples from each patient were separated by at least 2 weeks.
Phylogenetic Analyses
A direct cloning of PCR products was performed as described previously [Briat et al., 2005]. Clones were sequenced separately from each PCR amplification. Sequences were analyzed with Sequence Navigator software and aligned with ClustalX software [Thompson et al., 1997]. Gap-stripped sequence alignments were performed for each patient. Distances between pairs of sequences were estimated based on Kimura's two-parametric model of nucleotide substitution (DNAdist module—PHYLIP package, v3.67) [Felsenstein, 2001]. The means of all comparisons of pairwise genetic distances from each clone derived from rectal HIV DNA, from PBMC HIV DNA or from plasma HIV RNA to every other clone from the same tissue/fluid were calculated for each patient. Maximum-likelihood trees were constructed using the HKY model (PHYML v2.4.4) [Guindon and Gascuel, 2003]. The reliability of the branching orders was confirmed by performing 1,000 bootstrap resamplings. In order to confirm the topology of the resulting trees, the distance matrix was employed for phylogenetic analysis with the neighbor-joining method. Bootstrap values >70% were considered significant.
Statistical Considerations
The differences detected between pairwise distances among variants from the rectum, variants from PBMCs and variants from plasma were assessed using Mann–Whitney test.
RESULTS
Seven patients were included: patients A and B were at the time of the primary infection; patients C, D, E, and F at chronic infection, and patient G with AIDS (Table I). No confounding condition such as inflammation was observed during histopathological analyses of rectal tissue.
Overall, phylogenetic analyses were conducted on 522 sequences from clones derived from nine rectal samples, four plasma samples and nine PBMC samples (6–30 clones per sample, median: 25) (GenBank accession numbers: GQ859192–GQ859245 and GQ904239–GQ904706).
The phylogenetic analysis showed that all sequences of rectal clones and of blood clones from each patient clustered together, with no contamination between the samples from different patients (bootstrap = 100%) (data not shown).
Pairwise distances within HIV DNA quasispecies in the rectum, within HIV DNA variants in PBMCs and within HIV RNA variants in the plasma were calculated. These seven patients with diverse history of infection had very different levels of genetic diversity, with medians of pairwise distances within HIV DNA quasispecies in the rectum ranging from 0.1% to 2.2% depending on the patient taken into account (Fig. 1a), from 0.02% to 1.8% within HIV DNA quasispecies in the PBMCs (Fig. 1b) and from 0.04% to 1.95% within HIV RNA quasispecies in the plasma (data not shown).
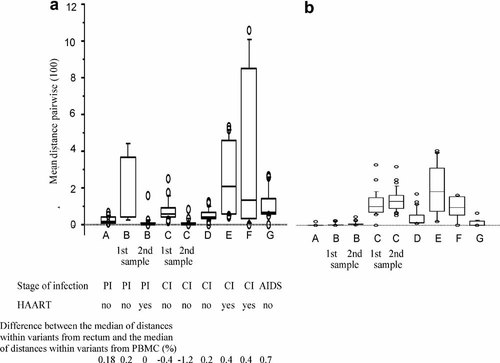
Distribution of pairwise distances among rectal HIV-DNA variants (a) and PBMC HIV-DNA variants (b). These boxplots represent the averages of comparisons of pairwise genetic distances from each clone derived from rectal HIV-DNA (a) or from PBMC HIV-DNA (b) to every other clone from the same tissue/fluid. Distances were computed by using the Kimura model and are displayed as 100 times the raw distance score. The horizontal lines show the median and the boxes comprise the first to third quartiles of calculated average genetic distances. A, B, C, D, E, F, and G concern viral clones from patient A, B, C, D, E, F, and G, respectively. Significant differences between pairwise rectum HIV-DNA distances and pairwise PBMC HIV-DNA distances were found for each patient (P < 0.01, Mann–Whitney test), except for patients under treatment (B: 2nd set of samples, E and F). PI, primary infection; CI, chronic infection; Patients A, B (1st sample), C are naïve to HAART; Patients D and G had received HAART but, at the time of sample collection, were not receiving treatment.
- (i)
In four out of five patients without HAART, the diversity among HIV DNA variants in the rectum was significantly greater than among HIV DNA variants in PBMCs (P < 0.0001). Differences between the median distance within variants in the rectum and the median distance within variants in PBMCs are presented in Figure 1 (patients A, B first set of samples, D, G). Surprisingly, this was observed from the primary infection stage (A, B first set of samples). The diversity among HIV DNA variants in the rectum was also significantly greater than among HIV RNA variants in the plasma (P < 0.0001). Differences between the medians distances within variants in the rectum and within variants in the plasma were +0.07%, +0.46%, +0.31% for the first set of samples of patients B, D and G, respectively.
- (ii)
During HAART, the genetic diversity within HIV DNA variants in the rectum and within HIV DNA variants in PBMCs for each patient was not statistically different (P = 0.14, 0.35, and 0.56 for patients B (second set of samples), E and F, respectively) (Fig. 1). It was also noticeable for patient B that the genetic diversity within HIV DNA quasispecies in the rectum was significantly higher before treatment (median 0.25%) than during HAART (median 0.12%) (P = 0.001).
Another objective was to check whether the HIV variants in the rectum and in the blood were interspersed or separated. Phylogenetic analyses showed that variants in the rectum and in the blood were interspersed partially or totally in all the seven patients (Fig. 2). In patients with sequential samples, most of the variants from the first set and variants in the blood from the second set were interspersed (B, C), showing the persistence of some archived variants in the body over time (Fig. 2). Nevertheless, clusters grouping at least some variants were observed in the rectum in patients A, B, C, E, F, G (bootstrap values >70%) (Fig. 2). It was also noticeable that all the clusters identified with the maximum-likelihood analysis were confirmed using a neighbor-joining approach.
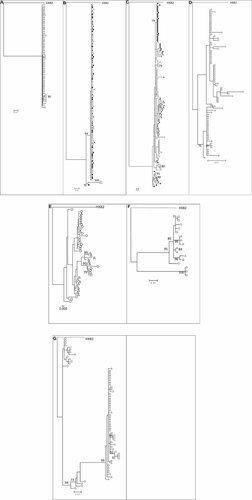
Phylogenetic relationship of viral clones derived from the blood and rectum (maximum-likelihood trees, using FR-HXB2 as root). The numbers near the nodes indicate the percentage of bootstrap replicates (1,000). Samples at inclusion: rectal HIV-DNA (open circle), blood HIV-DNA (open triangle), blood HIV-RNA (open square); second set of samples for two patients: rectal HIVDNA (full circle), blood HIV-DNA (full triangle), blood HIV-RNA (full square). A, B, C, D, E, F, and G illustrate viral clones from patient A, B, C, D, E, F and G, respectively. All the clusters identified with the maximum-likelihood approach were confirmed using a neighbor-joining analysis.
DISCUSSION
Studies carried out on the viral quasispecies in the different tissues could provide useful information enabling understanding of virus trafficking. Very little is known about the gut-associated lymphoid tissue in patients infected with HIV, due to the small size of samples and the difficulty in obtaining such samples. For the first time, the genetic diversity of HIV DNA in the rectum was compared to the genetic diversity of variants in blood, for well-characterized patients either at the time of the primary infection, or during chronic infection and or with AIDS. The HIV DNA variants include archived viral strains accumulated over time, as well as strains from recent infections; while the HIV RNA variants represent recently produced viral particles. Despite the low number of patients, statistically significant differences were observed.
These data suggest that variants in the rectum were more heterogeneous than variants in the blood in patients without HAART. To our knowledge, this is the first report to document that a viral genetic diversity seems to exist in the rectum from the time of the primary infection. For patient B (first set of data), the greater genetic diversity was mainly linked to two sequences of HIV DNA that clustered together. Unfortunately, other rectal HIV DNA sequences could not be recovered in patient B due to the low number of CD4+ T cells in relation to the small size of the rectal biopsies, and further studies will be necessary to confirm that point. One limitation of this study could be the bias introduced by the strategy of PCR cloning. However, a recent publication again discussed this point and concluded that this method of cloning is relevant if an adequate number of PCR templates is analyzed [Jordan et al., 2010]. In the present study, the number of PCR templates was high (Table I), therefore the strategy of PCR cloning could be considered with a limited bias. Although the existence of undetected minor variants which could influence the viral diversity in PBMCs and in the rectum cannot be excluded, the results may be explained by the activated milieu of the gut-associated lymphoid tissue which may provide an improved environment for the replication of HIV. In a larger group of patients [Avettand-Fenoel et al., 2008], we quantified the proportion of CD4+ rectal cells by immuno-staining in available biopsies for some patients and calculated the HIV DNA load per million of CD4+ rectal cells. It was noted that the HIV DNA load per million of CD4+ rectal cells was higher systematically than the HIV DNA level per million PBMCs, as described by Chun et al. [2008], Chomont et al. [2009], and Yukl et al. [2010]. The microenvironment in the gut-associated lymphoid tissue can be explained by the differences in the local immune selection pressures, the HIV-specific immune response, the constraints on viral entry and the replication rates due to the availability of activated target cells. The increased error prone replication would result in greater viral diversity.
These data suggest that heterogeneity was not significantly different in the rectum and in the blood during HAART, as described previously [Chun et al., 2008]. This observation suggests that HAART played a role in equalizing the dynamics of HIV in the gut-associated lymphoid tissue and PBMCs, although some clusters persisted.
In the cases described, the fact that the HIV variants in the rectum, in PBMCs and in the plasma were interspersed underlines that large exchanges of virus particles and/or infected cells occur between these tissues/fluids, as already shown in patients with HAART [Chun et al., 2008]. This is in line with the positive correlation observed between the HIV DNA levels in the rectum and in PBMCs at different stages [Avettand-Fenoel et al., 2008]. As observed previously with the variants of HIV in faeces [van der Hoek et al., 1998] and in the gut-associated lymphoid tissue [van Marle et al., 2010], complete, independent evolution of HIV-1 was not observed in the intestinal tissue.
Nevertheless, in most cases, at least some clones of HIV DNA in the rectum could be differentiated from variants in the blood, as described recently in the esophagus, the stomach, the duodenum, the colon and the blood with phylogenetic and drug mutation analysis [van Marle et al., 2010]. This suggests the existence of local replication foci which could play a major role in the immunopathogenesis of HIV-1 in the altered mucosa.
Finally, these results indicate exchange and trafficking of viral populations between blood and lymphoid rectal tissues. The results also suggest that viruses present in the rectum were more heterogeneous than variants in the blood in patients without HAART. Viral diversity is present in the rectum from the time of the primary infection. Studies on the gut-associated lymphoid tissue may improve understanding the pathogenesis of HIV, the assessment of treatment efficacy and the development of new therapeutic strategies, particularly those aiming at reducing the reservoir.
Acknowledgements
We are particularly indebted to the volunteers who agreed to participate in this project.
We thank Hayete Djarech who performed rectal biopsies with Eric Agoute, and we acknowledge the participants of the AC32 “Reservoirs” of the ANRS (Agence Nationale de Recherches sur le SIDA et les hépatites virales) for helpful discussions.




