Characterization of hepatitis B virus in turkish blood donors, and the prevalence of the SP1 splice variant
Abstract
Hepatitis B is a disease of the liver that can manifest acutely, or persist chronically as a result of infection with the hepatitis B virus (HBV). Turkey has a moderate endemicity level of HBV infection, and all data published to date has shown this to be of genotype D, predominantly of subgenotype D1. However the sequences of very few full genomes have been published. The aim of this study was to characterize the molecular profile of hepatitis B virus in asymptomatic, first-time Turkish blood donors. The results confirm that genotype D, subgenotype D1 is the most prevalent HBV strain in Turkey, accounting for 94% of cases. Subgenotypes D2 and D3 were present as minority strains (4% and 2%, respectively). A singly spliced HBV variant that is capable of forming defective HBV particles and has been associated with apoptosis and activation of T-cell responses was also detected in 52.5% of samples screened, co-circulating with wild type genomes. J. Med. Virol. 83:1321–1325, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Hepatitis B virus (HBV) carrier prevalence in Turkey is reported to be around 4% [Degertekin and Gunes, 2008], which is in line with other Mediterranean and Middle Eastern countries and meets the criteria for intermediate endemicity level. HBV belongs to the family Hepadnaviridae, and has a 3.2 kb genome that, when encapsidated within the virion, is a relaxed circular DNA molecule that is only partially double-stranded. It was first reported in 1989 that HBV could undergo splicing by the host's machinery in human hepatoma cells transfected with full-length HBV [Su et al., 1989]. This 2 kb variant is commonly termed SP1, and contains a 1223 nucleotide deletion beginning at the last codon of the core gene and spanning through to the middle of the S gene. To date, this splice variant has been identified in chronic hepatitis B patients infected with HBV genotypes A and D, and an additional 10 splice variants have been identified in patients infected with genotypes A, B, and D [Gunther et al., 1997].
HBV is divided into eight well-documented genotypes (A–H), some of which are further divided into two or more sub-genotypes [Kay and Zoulim, 2007]. Recently, two putative new genotypes, I and J, have been reported [Tran et al., 2008; Tatematsu et al., 2009]. Genotype D is predominant in the Mediterranean basin, and is also frequently found in populations from Europe, Africa, and Asia. Of the HBV genotypes, this is the least defined geographically, which may be a result of diverging earlier than the other genotypes [Norder et al., 2004]. The majority of the current data on HBV in Turkey has come from patients with clinical Hepatitis B, with few full genome sequences published. Sequencing of full genomes and the S gene from patients with chronic HBV infection identified only genotype D in Turkish samples, and where subgenotyping was documented, D1 was the most prevalent subgenotype, with a minority of subgenotype D2 strains present [Bozdayi et al., 2005; Serin et al., 2005; Sayiner et al., 2008; Sertoz et al., 2008]. The aim of this study was to characterize HBV strains obtained from HBsAg-positive plasma samples from asymptomatic, first-time, replacement blood donors collected in Ankara, Turkey, using serological and molecular techniques.
MATERIALS AND METHODS
HBsAg-positive plasma samples were collected during three separate periods in 2004, 2005, and 2006, with a total collection period of 13 months. Samples were heat-inactivated at 60°C for 30 min prior to shipping on dry ice, and upon arrival in the UK were stored at −80°C until ready to use. Viral DNA was extracted and quantified as described previously [Allain et al., 2003; Garmiri et al., 2009]. To generate sequence for the entire HBV genome from 50 samples, the Expand High Fidelity PCR system (Roche) was used for two nested PCR assays. Firstly, a nearly full-length fragment of approximately 3,000 bp, minus a 50 bp precore section was amplified as previously described [Zahn et al., 2008]. A second 300 bp fragment including the basic core promoter/precore (BCP/PC) 50 bp gap was produced to complete the full genome [Candotti et al., 2006]. PCR products were purified using the EZNA cycle pure kit (Omega, Lutterworth, UK).
A semi-nested PCR was used to screen for the SP1 variant, using newly designed primers located in the core and S genes. SP1_core (5′-GTCGCAGAAGATCTCAATCTCGGG-3′; position 2421–2444) was used as the sense primer in both first and second round PCR; SP1_R1 (5′-ATACAAAGGCATTAATGCAGGGTA-3′; position 1065–1042) and SP1_R2 (5′-TGTGTAAATGGAGCGGCAAAGCC-3′; position 1012–1034) were used as the anti-sense primers in first and second round PCR, respectively. Using the Roche Expand High Fidelity system, a 50 µl reaction mixture contained: 1× PCR buffer II (including MgCl2; final concentration 2.5 mM), 0.8 mM dNTPs, 0.3 µM each primer, and 5.25 U HiFi. For samples with a viral load of ×108 IU ml−1 or above, 3 µl of viral DNA was used as first-round template; 5 µl was used for samples with viral loads of ×104– × 107 IU ml−1 and 10 µl for samples × 103 IU ml−1 or below. Six microliters of the first-round reaction was loaded into the second-round PCR. An identical touchdown cycling program was used for both rounds, and consisted of: an initial denaturation step at 94°C for 5 min, followed by 10 cycles of 94°C/40 sec; 65°C/45 sec (reduced by 1°C each cycle); 72°C/1 min, and a following 30 cycles of 94°C/40 sec; 55°C/45 sec; 72°C/1 min. A final extension step was performed at 72°C for 10 min. In the presence of full-length HBV DNA (3.2 kb) a 1.8 kb product is amplified, in comparison to a 580 bp product for the SP1 variant, visualized using agarose gel electrophoresis.
Sequences with interesting deletions or insertions were examined further by fresh full genome or BCP/PC PCR reactions. Following addition of a 3′ single adenosine overhang to the PCR product, amplicons were ligated into either the pCR2.1 TOPO® TA vector (BCP/PC) or the TOPO® XL vector (Invitrogen, Paisley, UK) as per the manufacturer's instructions. Following transformation of chemically competent Escherichia coli cells, plasmid DNA was isolated using a QIAprep spin miniprep kit (Qiagen, Crawley, UK), and a minimum of 10 clones were sequenced.
Sequencing and phylogenetic analysis was performed using Seqman, SeqBuilder (DNASTAR), and MacVector software. PAUP* version 4.0 beta 10 was used to calculate genomic distances. Microsoft Excel and Prism (version 4) were used for analysis of HBV DNA and HBsAg load.
Antibodies against HBcAg (anti-HBc) and HBsAg (anti-HBs) were tested by ELISA with a Monolisa® anti-HBc PLUS kit (Bio-Rad, Hemel Hempstead, UK) and a Monolisa® anti-HBs PLUS kit (Bio-Rad, Marnes-la-Coquette, France), respectively.
RESULTS
Over the collection period samples were taken from 20,960 first-time, replacement blood donors (18,898 male: 2,062 female). Routine screening identified 200 samples as positive for HBsAg (187 male: 13 female), giving a prevalence of 1% for males and 0.6% for females. Plasma was obtained for 199 HBsAg positive samples, and quantification of HBV DNA load by Q-PCR was successful in 176 (88.4%) of these samples. The median viral load was 1.19 × 103 IU ml−1 (range 1.62 × 101–8.95 × 108 IU ml−1). In the remaining 23 samples (11.6%), HBV DNA was undetected by Q-PCR. However amplification of the BCP/PC region was achieved by nested-PCR in one of the 23 samples, meaning detection of HBV DNA was possible for a total of 177/199 samples (89%).
Complete genomic sequences were obtained for 50 samples (GenBank Accession numbers: JF754586–JF754635), from 57 randomly selected samples in which this was attempted. Phylogenetic analysis identified all strains as belonging to HBV genotype D; 47 clustered as subgenotype D1 (94%), 2 as subgenotype D2 (4%), and 1 as subgenotype D3 (2%). Sequence analysis of the S gene revealed the predominant serotype to be ayw2 (n = 41), 5 strains specified ayw3 and 1 strain was ayw4. An additional strain belonged to serotype adw, but could not be classified further, and serotype determination was not possible in one strain due to a deletion in the informative region (Table I).
| Feature | Results (no. of strains in which feature present/no. of strains screened) | ||||
|---|---|---|---|---|---|
| Genotype | D1 (47/50) | D2 (2/50) | D3 (1/50) | ||
| Serotype | adw (1/50) | ayw2 (41/50) | ayw3 (5/50) | ayw4 (1/50) | Unknown (2/50) |
| X ORF | Deletions (0/50) | Insertions (1/50) (6 bp) | Truncations (0/50) | ||
| Pol ORF | Deletions (6/50) (6–66 bp) | Insertions (1/50) (6 bp) | Truncations (0/50) | ||
| Pre-S/S ORF | Deletions (6/50) (6–66 bp) | Insertions (0/50) | Truncations (4/50) (12–474 bp) | ||
| Pre-C/C ORF | Deletions (1/50) (37 bp) | Insertions (2/50) (1–2 bp) | Truncations (2/50) (369–417 bp) | G1896A (33/50) | |
| Viral load (VL) (IU/ml) | Undetectable (23/199) | <100 (26/199) | 1 × 102–9 × 103 (120/199) | 1 × 104–9 × 106 (22/199) | >1 × 107 (8/199) |
| Splice variant (SP1) frequency | VL < × 104 (3/12) | VL ≥ × 104–< × 106 (8/18) | VL > × 106 (10/10) | ||
- Summary of the features identified in the HBV strains sequenced, including deletions, truncations, and insertions in the four ORFs, and viral load and SP1 distribution (HBV, hepatitis B virus).
PreC/C
One sample contained a deletion of 37 nucleotides, resulting in a frameshift mutation and premature protein truncation. In another strain, insertion of two adenine nucleotides in the precore region caused a frameshift mutation, which is predicted to result in the inclusion of 31 amino acids in the precore protein, whilst leaving the core protein unaffected. Cloning of the BCP/PC for this strain identified the double adenine insertion in all 10 clones sequenced. Serum testing of these two samples identified the presence of anti-HBc. Another sample contained a single adenine insertion within the core region, introducing a premature stop codon and protein truncation. Serum was not available for anti-HBc testing of this sample. Seven samples (14%) contained mutations that abolished the precore initiation codon. The G1896A mutation, which introduces a premature stop codon and abolishes precore protein and HBeAg synthesis, was present in 33 strains (66%).
Pre-S/S
Mutations disrupting the initiation codon of Pre-S2 were identified in three samples (6%). Five samples contained deletions in the Pre-S/S transcript, ranging in size from 6 to 51 nucleotides. These were present throughout the three ORFs. Three samples contained nonsense mutations that are predicted to result in premature truncation of the protein. One further sample contained a 66 nucleotide deletion spanning the Pre-S1/Pre-S2 boundary, in addition to a nonsense mutation leading to premature truncation of the S protein at amino acid residue 69. Testing for the presence of anti-HBs in the serum from which this strain was obtained was negative. Cloning of this sample identified the presence of wild-type species, in addition to a much smaller product with a 1223 nucleotide deletion (positions 2449–489), predicted to result from splicing of viral mRNA. This deletion started at the final codon of the core gene and extended into the middle of the S gene.
The mean overall amino acid divergence within the Pre-S/S region was 2.7% (range 0–6.9%). The major hydrophilic region (positions 100–169) was more variable, with a mean amino acid substitution rate of 5.3% (range 0–20%). No substitutions associated with vaccine-escape mutants were identified in this cohort.
X Gene
No deletions were present within the X transcript in any samples. However, one strain contained a 6 nucleotide insertion, leading to the inclusion of two additional amino acid residues (glycine and methionine) in the X protein, inserted between wild-type residues 79 and 80. The basal core promoter (BCP) variants, A1762T and G1764A were present as a dual mutation in 10 strains, with an additional six samples carrying G1764A as a single change.
Pol
Six strains carrying deletions in the polymerase gene ranging between 6 and 66 nucleotides were identified; these were all located in the spacer domain of the protein. There were no disruptions to the highly conserved YMDD motif in any of the 50 strains sequenced. One further sample had a 6 nucleotide insertion within the RNase H domain of the protein.
SP1 Screening
Screening for the presence of the SP1 variant was successful in 40/43 samples in which this was attempted (Fig. 1). A full genome sequence was available for 27 of the 40 successfully amplified samples. The SP1 variant was present in 10/10 (100%) samples with viral loads ≥106, in 8/18 (44%) samples with viral loads of ≥104–<106, and in 3/12 (25%) of samples with viral loads <104.
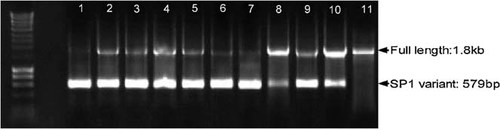
Representative gel of SP1 screening. Amplification of full-length sequence results in a 1802 bp product, whereas spliced HBV DNA, which contains a 1223 nucleotide deletion, generates a 579 bp product. Depending on the ratios present, both products may be amplified or alternatively only one amplicon is generated. Sample 1: TK89; Sample 2: TK88 (JF754601); Sample 3: TK129 (JF754618); Sample 4: TK32; Sample 5: TK28 (JF754624); Sample 6: TK74; Sample 7: TK171 (JF754623); Sample 8: TK178; Sample 9: TK63 (JF754608); Sample 10: TK38 (JF754591); Sample 11: (TK70). (Marker: Bioline, Hyperladder I; HBV: hepatitis B virus.)
DISCUSSION
Analysis of 50 full HBV genotypes extracted from asymptomatic Turkish blood donors selected at random supports previous findings that the most prevalent HBV strain in this country is subgenotype D1 and serotype ayw2 [Bozdayi et al., 2005; Sertoz et al., 2008].
In previously published complete HBV genomes from chronic Hepatitis B patients from Turkey, the G1896A variant was identified in 18% of samples, and these patients were HBeAg negative [Bozdayi et al., 2005]. In this cohort from healthy volunteer donors, the G1896A precore stop variant was identified in 66% of samples. This is in keeping with data that shows a high prevalence of the G1896A mutation in samples from Mediterranean populations [Funk et al., 2002]. G1896A has previously been documented to be present in 15% of HBeAg-positive Turkish patients, compared to 85% of HBeAg-negative patients [Bozdayi et al., 1999, 2005]. This mutation is of interest as it results in the prevention of HBeAg synthesis, and seroconversion of HBeAg to anti-HBe is correlated with a decline in viral load [Tedder et al., 2002]. The core promoter dual mutations (A1762T, G1764A) were identified previously in 3/11 chronic-HBV infected patients, one of whom was HBeAg positive [Bozdayi et al., 2005], and was identified in 20% of the strains sequenced in this study. The double mutation has been implicated with a reduction in the generation of precore mRNA and HBeAg production, and is more prevalent in patients with advanced liver disease [Poustchi et al., 2008]. Given the relatively high frequency of these mutations in this study population, and their clinical implications, it may be of use to sequence the viral genome in subjects testing positive for HBV-infection in Turkey.
This is the first time that screening of the SP1 variant has been conducted in asymptomatic blood donors. The data demonstrated that the SP1 variant was present in more than half of the samples screened (21/40), and was ubiquitous in samples with viral loads ≥106 IU/mL. However, that SP1 is detected at a much lower frequency in samples with low viral loads (3/10 VL < 104 IU/ml) may not be directly linked to viral load, but may be a matter of the ratio at which the transcripts are present and the ability of the PCR to detect very low levels of the spliced variant. Previous studies have examined the prevalence in symptomatic HBV-infected patients, and have correlated its presence with chronic infection and hepatocellular carcinoma [Lin et al., 2002]. The association between prevalence of the HBV splice variant and severity of liver disease is not yet elucidated; one study reported an association between a high proportion of spliced HBV: wild-type HBV in patients with more severe liver necroinflammation [Soussan et al., 2008], whereas a different study found no association between spliced HBV prevalence and disease status [Preiss et al., 2008]. Decreased levels of SP1 have been observed in patients with a lamivudine-resistant HBV strain, suggesting that antiviral therapy disrupts the balance between circulating HBV splice variants and full-length HBV genomes [Preiss et al., 2008].
In addition, a novel 10.4 kDa protein (HBV splice-generated protein, HBSP) arose from the fusion of a section of the polymerase and a new ORF created downstream of the splicing event [Soussan et al., 2000]. Detection by Western blot analysis has revealed the presence of this protein in 46% of serum samples from chronic HBV carriers, as opposed to <1% of healthy HBV-negative controls [Soussan et al., 2003]. Functionally, this protein has been demonstrated in vitro to increase hepatocyte apoptosis [Soussan et al., 2000], and in humans and transgenic mice to induce a T-cell response [Mancini-Bourgine et al., 2007].
Acknowledgements
The authors would like to thank Mrs Yeşim Özer and the staff of the Blood Bank & Apheresis Unit, Ankara, Turkey who participated in the collection and screening of blood donor samples, in addition to Dr. B.H.M. Meldal and Dr. I.H.A. Barnes for their contribution to DNA extraction and quantification.




