Abstract
Human parechoviruses (HPeVs) are widespread pathogens belonging to the Picornavirus family. The aim of the present study was to assess the prevalence and genetic diversity of HPeV in Shanghai, China, during a HPeV screening program in 2008 and 2009. Of 300 stool samples from children under the age of 5 years with acute diarrhea seen at Children's Hospital, Fudan University, Shanghai, China, 165 (55%) were HPeV-positive. The median age of infected children was 3 months. The prevalence of HPeV was high (57%) in infants up to 2 years old but dropped to 30.4% in children between 2 and 5 years old. The prevalence did not differ by sex. Infections were present throughout the year but peaked in July and August. The most predominant genotype was HPeV1. Of the 139 strains, 4 were found in 9 samples: HPeV4 (n = 4), HPeV5 (n = 1), HPeV6 (n = 1), and HPeV8 (n = 3). This study provided useful data on the epidemiology of HPeV infection by documenting the distribution of genotypes, age of infection, and seasonal patterns in Shanghai, China. J. Med. Virol. 83:1428–1434, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Human parechoviruses (HPeVs) are members of the large and growing family of Picornaviridae. Two types, HPeV1 and HPeV2, initially known as echovirus 22 and 23 (EV22 and EV23), were first isolated in 1956 in the USA from stool specimens of children with diarrhea [Hyypiä et al., 1992]. Although the majority of HPeV infections occur early in life without specific symptoms, disease manifestations associated with many of the types have been described, ranging from gastroenteritis and respiratory infections to neurological disease, particularly in neonates [Ito et al., 2004; Boivin et al., 2005; Benschop et al., 2006a; Baumgarte et al., 2008; Harvala et al., 2008; Harvala and Simmonds, 2009].
The 14 known HPeV genotypes were discovered mainly in young children, but complete genome sequences are only available for HPeV types 1 through 8. Infections with HPeVs are widespread, but different HPeV genotypes have different epidemiological and clinical characteristics.
HPeV1 is a widely dispersed pathogen that mainly affects young children [Joki-Korpela and Hyypiä, 2001; Legay et al., 2002; Abed and Boivin, 2006; Zhang et al., 2009; Benschop et al., 2010; Ito et al., 2010; Pham et al., 2010a, 2011a,b]. HPeV2 infections are rare and are mostly associated with gastrointestinal symptoms [Ehrnst and Eriksson, 1996]. HPeV3 was identified in Japan from a 1-year-old child with transient paralysis, fever, and diarrhea in 1999 [Ito et al., 2004], and the prevalence may now be either equal to HPeV1 or a close second [Joki-Korpela and Hyypiä, 2001; Legay et al., 2002; Abed and Boivin, 2006; Zhang et al., 2009; Benschop et al., 2010]. HPeV3 infection causes more severe diseases, such as sepsis or sepsis-like illness and meningitis or encephalitis in relatively young children [Ito et al., 2004; Boivin et al., 2005; Abed and Boivin, 2006; Benschop et al., 2006a; Watanabe et al., 2007; de Vries et al., 2008; Wolthers et al., 2008; Harvala et al., 2009; Levorson et al., 2009; Selvarangan et al., 2011]. Symptoms of central nervous system (CNS) involvement, as with encephalitis and paralysis, have been reported for HPeV1 as well [Figueroa et al., 1989; Koskiniemi et al., 1989; Legay et al., 2002; Selvarangan et al., 2011], but less frequently than in HPeV3 infections [Abed and Boivin, 2006; Benschop et al., 2006a; Selvarangan et al., 2011].
HPeV4 through 8, 10, 11, and 14 were discovered, respectively, in the Netherlands, the United States, Japan, Pakistan, Brazil, Sri Lanka, and Thailand [Oberste et al., 1998; Benschop et al., 2006b, 2008; Watanabe et al., 2007; Drexler et al., 2009; Li et al., 2009; Calvert et al., 2010; Pham et al., 2010b, 2011a]. Finally, there are unpublished reports of types 9, 12, and 13 (http://www.picornastudygroup.com/types/parechovirus/hpev.htm) based on VP1 sequence comparisons [Harvala and Simmonds, 2009], but clinical data on these types are lacking.
The aim of this study was to gain insight into the prevalence and genetic diversity of HPeV in Shanghai, China, by direct screening and typing of stool samples from children with acute diarrhea under the age of 5 years in 2008 and 2009.
MATERIALS AND METHODS
Sample Collection
From 1 January 2008 to 31 December 2009, a total of 300 rotavirus-negative stool specimens were collected from children with diarrhea (<5 years old) who were hospitalized at Children's Hospital, Fudan University, Shanghai, China. Because the stool specimens were collected in the normal course of patient care, neither Institutional Review Board approval nor informed consent was required for this study.
Parechovirus Testing
From supernatants of 20% (w/v) of stool specimens in saline, RNA was extracted by TRIzol (Invitrogen, Carlsbad, CA), according to the manufacture's instructions. The RNA was dissolved in 10 µl of RNase-free water. Reverse transcriptase reaction (PrimeScript™ RT kit, Takara, Dalian, China) was performed in a reaction volume of 10 µl according to the manufacturers' instructions using 4 µl of the extracted RNA, 100 µmol of random primers, and 2.5 U of reverse transcriptase. The reaction mixture was incubated at 37°C for 30 min and then heated to 85°C for 5 sec. cDNA was either used directly for PCR or stored at −20°C.
Testing for HPeV was carried out using the highly conserved 5′UTR primers [Harvala et al., 2008]. In the first PCR step, 2 µl of the initial RT reaction product was added to a PCR reaction mixture containing 0.5 µmol each of outer primers, 50 µmol of dNTP, and 0.625 U of Ex Taq DNA polymerase (Takara) in 25 µl of volume.
The PCR cycling conditions were 40 cycles: 94°C for 30 sec; 55°C for 30 sec; 72°C for 30 sec; and a final incubation of 72°C for 7 min. One microliter from the first round of PCR was used as the template in a second round of PCR with inner primers and amplified using the same cycling conditions. PCR products were run on 2% agarose gels prestained with Golden view to identify positive samples with a predicted size of 243 bp.
Parechovirus Genotyping
For parechovirus genotyping, samples testing positive by the 5′UTR were amplified by nested PCR using primers from the VP3/VP1 junction region as described elsewhere [Harvala et al., 2008]. Four microliters of cDNA was used for the first round of PCR, which was performed in a 25-µl volume containing 0.5 µmol each of outer primers, 50 µmol of dNTP, and 0.625 U of Ex Taq DNA polymerase using the following cycling conditions: 40 cycles of 94°C for 30 sec; 50°C for 30 sec; 72°C for 30 sec; and a final incubation of 72°C for 7 min. Then, 2.5 µl of the product was transferred for a second round of PCR with 1 µmol each of VP3/VP1 inner primers, 100 µmol of dNTP, and 1.25 U of Ex Taq DNA polymerase in a final volume of 50 µl using the same PCR cycling conditions. The final product of 304 bp was visualized by agarose gel electrophoresis as described above.
DNA fragments of 304 bp were purified from agarose gels using a QIAquick Gel Extraction kit (Qiagen, Hilden, Germany). The nucleotide sequence of each PCR product was determined using the BigDye sequencing kit on a 3730 sequencer (Pekin-Elmer Applied Biosystems, Foster City, CA). Sequence analysis of the PCR product of each strain was analyzed with Seqscanner (https://products.appliedbiosystems.com), and genetic identity was determined by comparing the sequence with standard strains in Genbank. A multiple-sequence alignment was constructed using ClustalW, and phylogenetic trees were constructed using Mega 4.1 software by the neighbor-joining methods [Thompson et al., 1994; Tamura et al., 2007].
Statistical Methods
Data were analyzed with SPSS version 14 for Windows. The prevalence of HPeV infection between boys and girls was compared with χ2 test. Alpha was set at P < 0.05, and all tests were two-tailed.
RESULTS
Between January 1, 2008 and December 31, 2009, 300 rotavirus-negative stool specimens were collected from 300 children. The majority of the children had been born at term (285/300, 95%). All children had diarrhea during hospitalization and more than half (167/300, 55.7%) had respiratory symptoms. Fever was present in 91% and a rash was noted in 17 (17/300, 5.6%). More than 10% of children (31/300, 10.3%) had CNS symptoms and 14 (14/300, 4.6%) had received a diagnosis of sepsis-like illness. Another 25 children had other symptoms, such as the signs of neonatal infections, urinary tract infection, measles, or cardiac arrhythmia.
Age and Seasonal Distributions of Parechovirus Infection
Of the 300 samples tested for HPeV, 165 (165/300, 55%) were positive. The prevalence of HPeV infection did not differ significantly between boys and girls (54.5% vs. 55.9%, P > 0.1). The median age of infected children was 3 months.
The frequency of HPeV infection was 59.7% (40/67) in neonates, 57.1% (56/98) in infants from 1 to 5 months old, 56.7%(34/60) in small children of 5–11 months old and it remained high (53.8%, 28/52) in children of 12–23 months old. Prevalence of HPeV declined to 30.4% (7/23) in children of 24–60 months old (Fig. 1).
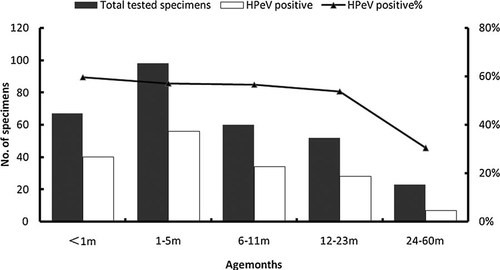
Prevalence of human parechovirus infections in 165 hospitalized children in Shanghai, China, 2008–2009, by age group. Bars denote the number of detected cases in different age groups and lines indicate the positive rate of HPeV infections in each age group.
HPeV infections were present through the year. Circulation of HPeV was low in January and February, then it increased gradually and peaked in summer and autumn, with high prevalence in July (75%, 21/28) and August (87.1%, 27/31) (Fig. 2).
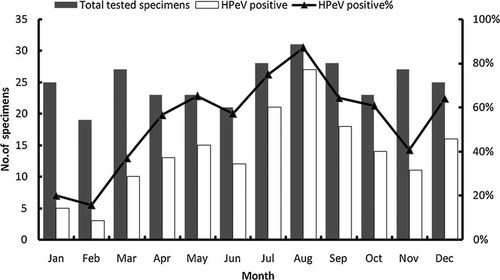
Seasonal distribution of HPeV infections among children hospitalized in Shanghai, China, 2008–2009. Bars represented the number of detected cases and the positive rate of HPeV infections in each month was indicated in lines.
Prevalence of Parechovirus Genotypes
All 165 HPeV-positive samples were analyzed by Mega 4.1. Genotyping was successful in 139 (84%) samples. The most predominant genotype detected over both years was HPeV1 (92.1%, 128/139). Only four samples were identified as HPeV4 (2.9%, 4/139); one sample was HPeV5; and one was HPeV6 (0.7%, 1/139). All six genotypes were detected only in 2009 (Table I). In addition, three samples (2.2%, 3/139) were identified as HPeV8 in 2008. Although HPeV1 was present through the year, only HPeV4, HPeV5, and HPeV6 were found from July to October, and HPeV8 was only found in May.
| Year | Number of children testing positive for parechovirus | Total | |||||
|---|---|---|---|---|---|---|---|
| HPeV1 | HPeV4 | HPeV5 | HPeV6 | HPeV8 | Untyped | ||
| 2008 | 65 | 0 | 0 | 0 | 3 | 2 | 70 |
| 2009 | 63 | 4 | 1 | 1 | 0 | 0 | 69 |
| Total | 128 | 4 | 1 | 1 | 3 | 2 | 139 |
The phylogenetic relationship of the sequences of detected in the 139 HPeV samples was compared to 36 references from Genbank. The phylogenetic tree was generated for 304 nt from region VP3/VP1 of selected HPeV strains (Fig. 3). The largest cluster was HPeV1. The majority HPeV1 strains shared only 75–82% identity with Harris, the prototype strain. Seven strains clustered closely with the Harris strain (89% nucleotide identity). Otherwise, there was no specific temporal separation among the different HPeV1 strains. The HPeV4 strains had 85–87% nucleotide identity with the reference strain T75-4077, 84–86% with K251176-02, and 87–89% with Fuk2005-123. The nucleotide identity of HPeV5 strain was 82% for CT86-6760 and 80% for T92-15. The HPeV6 strain had 89% nucleotide identity with NII561-2000, 88% with BNI-67/03, and 90% with 2005-823. HPeV8 strains and BR/217/2006 had 81% nucleotide identity.
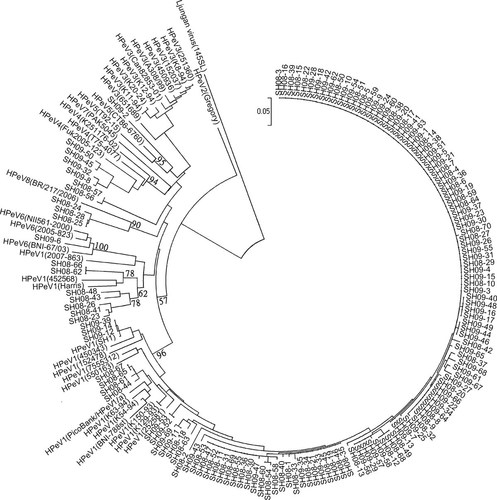
Rooted phylogenetic tree base on amino acid differences in the VP3/VP1. Reference strains of different HPeV genotypes were obtained from NCBI Genbank. HPeV1 reference strains and isolates (accession numbers are in parentheses), are Harris (S45208); PicoBank/HPeV1/a (FM242866); BNI-788st (EF051629); SH1 (FJ840477); 7555312 (FM178558); 152478 (GQ183018); 252581 (GQ183019); 450343 (GQ183020); 550163 (GQ183021); K129-93 (GQ183022); K150-93 (GQ183023); K54-94 (GQ183024); K63-94 (GQ183025); 2007-863 (GQ183034); 452568 (GQ183035); the HPeV2 strain is Gregory (AF055846); the HPeV3 strains and isolates are Can82853-01 (AJ889918); A308/99 (AB084913); 152037 (GQ183026); 251360 (GQ183027); 450936 (GQ183028); 651689 (GQ183029); K11-94 (GQ183030); K12-94 (GQ183031); K20-94 (GQ183032); K8-94 (GQ183033); the HPeV4 strains and isolates are T75-4077 (AM235750); Fuk2005-123 (AB433629); K251176-02 (DQ315670); the HPeV5 strains and isolates are CT86-6760 (AJ005695); T92-15 (AM235749); the HPeV6 strains and isolates are NII561-2000 (AB252582); BNI-67/03 (EU024629); 2005-823 (EU077518); the HPeV7 strain is PAK5045 (EU556224); and the HPeV8 strain is BR/217/2006 (EU716175). Ljungan virus 145SL (AF327922) was used as an outgroup.
However, two positive HPeV samples obtained in 2008 could not be assigned to be a specific HPeV type. The two strains have 98% nucleotide identity. Strain SH08-168 had the best nucleotide identity, 79% to the prototype strain HPeV5 CT86-6760. The second-best match was 78% identity with HPeV4 T75-4077.
Characterization of Clinical Symptoms
Diarrhea was present in all children with HPeV, and fever was recorded in 95%. More than half (55.2%, 91/165) had signs of respiratory tract illness, and 10 (6.1%, 10/165) received a diagnosis of sepsis-like illness. Meningitis was diagnosed in 2 children (1.2%, 2/165), and encephalitis with seizures was diagnosed in 12 (7.3%, 12/165).
Other clinical symptoms were those of hand foot and mouth disease (1.8%, 3/165), neonatal infections (3.6%, 6/165), urinary tract infection (1.2%, 2/165), measles (1.8%, 3/165), cardiac arrhythmia (1.2%, 2/165), and rash (3%, 5/165).
DISCUSSION
In the 2-year study period, a high frequency (55%) of HPeV infections was detected in hospitalized children <5 years old by direct nested RT-PCR screening from stool specimens. However, the results here cannot be compared to those of other studies [Abed and Boivin, 2006; Watanabe et al., 2007; Baumgarte et al., 2008; Benschop et al., 2008; van der Sanden et al., 2008; Drexler et al., 2009; Ito et al., 2010; Pham et al., 2010a, 2011a,b] because these other studies were potentially biased from: (1) being based primarily on culture isolates, which can be biased because of the inability of the virus variants to replicate in certain cell lines; (2) being based one-step PCR directly screened from stool samples; or (3) the samples excluded the other gastroenteritis-associated viruses, such as rotavirus, human calicivirus, astrovirus, adenovirus, norovirus, and human bocavirus. The inconsistencies are unlikely to be caused by cross-contaminations with strict control measures in nucleic acid extraction and PCR analysis.
A longitudinal Norwegian study determined that 43% of 102 infants had had at least one HPeV infection by 12 months of age, whereas 86% had encountered the virus by age 2 years [Tapia et al., 2008]. The high prevalence of HPeV is consistent with serological studies showing that about 90% of children have been infected with at least one HPeV type by the age of 2 years [Joki-Korpela and Hyypiä, 1998; Takao et al., 2001; Ito et al., 2004; Abed et al., 2007; Tauriainen et al., 2007]. In addition, a recent case–control study from Lanzhou, China, showed that cases co-infected with HPeV and other gastroenteritis-associated viruses did not differ significantly from controls in either viral load or the frequency of HPeV infection [Zhang et al., 2010]. These authors suggested that HPeV is an “innocent bystander” that does not contribute to gastroenteritis. In this respect, asymptomatic carriage may be another reason the HPeV infection rate was high in the present study.
The percentage of HPeV-positive children was 57% (158/277) in children <2 years old, but the prevalence decreased to 30.4% after age 2 years. This age distribution is consistent with those of other studies, which found that HPeV infection was mostly restricted to children under 2 years of age [Abed and Boivin, 2006; Tauriainen et al., 2007; Harvala et al., 2010; Pham et al., 2011a].
In Shanghai, HPeV infection had a distinct seasonal distribution that peaked during July, August, and September. However, different seasonal patterns have been observed in other regions. Some studies reported a higher frequency in autumn and winter in Western Europe [Benschop et al., 2008; Tapia et al., 2008; van der Sanden et al., 2008]. A study in Japan demonstrated HPeV infection peaked in February [Pham et al., 2011b].
The HPeV1 genotype was the only predominant genotype during 2008 and 2009 in Shanghai, which is consistent with the fact that HPeV1 is the major genotype worldwide [Watanabe et al., 2007; Baumgarte et al., 2008; Benschop et al., 2008, 2010; Harvala et al., 2008; Tapia et al., 2008; Boros et al., 2010; Ito et al., 2010; Pham et al., 2010a, 2011a,b; Zhang et al., 2010]. HPeV1 strains could be grouped into two clusters, of which one comprises Harris-like variants designated clade 1A and another is designated clade 1B [Benschop et al., 2009]. As the prototype HPeV1 strain, Harris has been identified rarely in recent years [Benschop et al., 2008, 2009]. The fact that seven strains were found as Harris in the year of 2008 in this study shows that “old ”strains were still circulating in some of years, albeit at low frequency.
However, testing did not detect HPeV3, the second most prevalent genotype in many studies [Joki-Korpela and Hyypiä, 2001; Legay et al., 2002; Abed and Boivin, 2006; Zhang et al., 2009; Benschop et al., 2010; Ito et al., 2010; Pham et al., 2011b]. In Europe, HPeV3 shows a distinct cycle of infection, occurring almost exclusively in even-numbered years since 1988 [Benschop et al., 2006a, 2008; van der Sanden et al., 2008; Harvala et al., 2009]. Because HPeV3 was identified in Lanzhou in 2006 and 2007 [Zhang et al., 2010], it is speculated that 2008 and 2009 may not have been the prevalent seasons in Shanghai. More study and data are needed to confirm a periodical occurrence for HPeV3 in China.
In 2009, HPeV types 4 through 6 were identified in six HPeV-positive children. These types have only recently been discovered, and they are less common globally than types 1 and 3 [Baumgarte et al., 2008; Benschop et al., 2008; Harvala et al., 2008; van der Sanden et al., 2008; Pajkrt et al., 2009; Zhang et al., 2010]. All six of these children showed only gastrointestinal or respiratory symptoms, as did those in a Dutch report of relative young, symptomatic children infected with HPeV4 through 6 infections and who had respiratory symptoms, gastrointestinal symptoms, or both [Pajkrt et al., 2009]. In 2008, 4.3% (3/70) of the positive samples in the present study were HPeV8, and these three children also showed only signs of gastrointestinal or respiratory infection. Additional studies are needed to determine the characteristic and clinical features of HPeV4–8 infections in China.
This study provides useful epidemiological data on the features of HPeV infection by documenting the genotypes distribution, as well as age and seasonal patterns, in Shanghai, China.




