Evidence for positive selection in the extracellular domain of human cytomegalovirus encoded G protein-coupled receptor US28
Abstract
The human cytomegalovirus (HCMV)-encoded chemokine receptor US28 is also a seven-transmembrane G-protein coupled receptor, whose signaling pathway is known for its involvement in host immune system evasion. HCMV infection can result in serious disease in immunocompromised individuals and is also linked to atherosclerosis and cardiovascular disease. Identifying amino acid residues that play a crucial role in successful viral adaptation in response to the host's immune defense is critical for effective drug design. In this study maximum likelihood-based codon substitution analyses were carried out to determine whether any codon of US28 has evolved adaptively. If the rate of nonsynonymous (dn) to the rate of synonymous (ds) nucleotide substitutions (ω = dn/ds) is greater than one, the codon is said to be under positive selection, indicating adaptive evolution. Although the overall ω for US28 gene was 0.154, indicating that most codon sites were subject to strong purifying selection, five codon sites are under strong positive selection. Three (E18D/L, D19A/E/G, and R267K/Q) of these positively selected sites are located in extracellular domains, the domains that play a crucial role for successful viral adaptation in response to the host's immune defense. The C-terminal (R329Q/W) and the fifth transmembrane domain (V190I), each have one positively selected site. These results suggest that relative to the extracellular domains, amino acid residues present in intracellular domains are more selectively constrained. A few amino acid residues in extracellular domains of US28 evolved more rapidly, presumably due to positive selection pressure resulting from ligand-binding and pathogen interactions of extracellular domains. J. Med. Virol. 83:1255–1261, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Human cytomegalovirus (HCMV/HHV-5), a β-herpesvirus, is highly prevalent in general populations, affecting 30–90% of individuals in developed countries [Staras et al., 2006]. HCMV infection is asymptomatic in immunocompetent individuals but can result in serious disease in immunocompromised individuals [Soderberg-Naucler, 2006] and congenitally infected infants [Ross and Boppana, 2005]. Furthermore, HCMV infection is linked to several pathologies such as atherosclerosis [Hsich et al., 2001], cardiovascular disease [Muhlestein et al., 2000], and specific human malignancies [Cinatl et al., 2004; Soderberg-Naucler, 2006].
The 235 kb genome of HCMV contains more than 200 genes, including the G-protein coupled receptors US27, US28, UL33, and UL78. US28 is closely related to receptors for β-chemokines and binds a broad spectrum of chemokines, including CCL2/MCP-1, CCL5/RANTES, and CX3CL1/Fraktalkine. Although chemokine ligands interact with multiple sites on the extracellular face of chemokine receptors, the N-terminal 22 amino acids of US28 are required for binding of all chemokines [Casarosa et al., 2003]. In particular, a hexapeptide sequence (a 6 aa segment between amino acids 11 and 16) in the N-terminus of US28 has been shown to be critical for high-affinity binding of chemokine ligands [Casarosa et al., 2005]. In accordance with the chemokine binding profile, the US28 receptor has been suggested to act as a chemokine sink by binding and withdrawing chemokines from the HCMV-infected cells, thereby allowing them to escape host immune surveillance and spread in the host system [Bodaghi et al., 1998; Kledal et al., 1998; Pleskoff et al., 1998]. The intracellular carboxy-terminal domain of US28 has been identified as an important regulator of US28 signaling [Mokros et al., 2002; Miller et al., 2003; Waldhoer et al., 2003]. Deletion of the C-terminal 40 amino acids of US28 results in a more potent phospholipase C-β signal than wild-type US28, and a prolonged calcium signal in response to CCL5, suggesting that the US28 carboxy-terminal domain plays an important role in regulating agonist-independent or dependent signaling in infected cells [Stropes et al., 2009].
Previous studies have reported amino acid polymorphisms among the US28 genes of clinical isolates from children and adults [Arav-Boger et al., 2002; Goffard et al., 2006; Xia et al., 2006]. The comparison of US28 from AIDS patients with the sequences from uninfected children has shown a specific mutational profile [Xia et al., 2006]. Depending on the location of the substitution and the nature of substitutions (whether synonymous or nonsynonymous), nucleotide polymorphisms in US28 coding for chemokine receptors can either have a significant effect on receptor activity or little effect. If a nucleotide substitution alters the amino acid then it is referred to as a nonsynonymous (dn) substitution, if the nucleotide substitution does not alter the amino acid then it is referred to as a silent/synonymous (ds) substitution. The ratio of amino acid replacement to the silent substitution (ω = dn/ds) is the measure of selection pressure on a protein coding gene. When ω = 1, or ω < 1 or ω > 1, the protein coding gene is said to be under neutral, purifying, or positive selection, respectively. Amino acid substitutions resulting in alterations at key ligand binding extracellular domains might result in disrupted or abnormal receptor activity [Fernandez and Lolis, 2002; Metzger and Thomas, 2010]. Because of their crucial role in signaling immune responses, chemokine receptors might be subject to intense selection to accommodate signaling molecules, and thus are expected to experience purifying selection to maintain conformation and the functions of ligand binding and signaling [Kunstman et al., 2003; Metzger and Thomas, 2010]. However, chemokine receptors are also expected to experience positive selection pressure for successful viral adaptation in response to the host's immune defense. Therefore, receptors that are responsible for successful viral adaptation may experience balancing selection, which may result in the maintenance of polymorphism [Hedrick, 2007]. Identifying codons that are under positive selection and mapping the locations of these positively selected sites could be helpful in identifying targets of the immune response and hence help in vaccine design [de Oliveira et al., 2004].
Given this evolutionary trade off, it is of interest to investigate the signature of selection on the US28 chemokine receptor. Using the complete US28 sequences representing different geographic regions (USA, Germany, UK, France, and China) and infection status (HIV positive or negative patients), the present study reports on the degree of geographic association of clinical samples and the pattern of genetic polymorphisms, as well as the underlying genetic mechanism that maintains the genetic polymorphisms.
MATERIALS AND METHODS
Phylogenetic Analysis
A total of 103 complete US28 nucleotide coding sequences of human cytomegalovirus were retrieved from GenBank. These sequences were isolated from laboratory and clinical samples (HIV positive or negative patients), representing different geographic regions (USA, China, France, Germany, and UK). The coding sequences were aligned using MEGA4.1 [Tamura et al., 2007]. To infer phylogenetic relatedness among these isolates, a maximum likelihood (ML) tree was constructed with the appropriate model of nucleotide substitutions selected by Akaike Information Criterion (AIC) as implemented in ModelTest ver 3.7 [Posada and Crandall, 1998]. The ML tree was reconstructed using the heuristic search option, implementing stepwise addition with 100 random addition replicates and tree bisection-reconnection branch swapping in PAUP* version 4beta10 [Swofford, 2002]. Prior to selection analyses, a recombination detection program (RDP) implemented in the RDP3 software package [Martin et al., 2005] was used to reveal any evidence of recombination. The analyses revealed no evidence of recombination, thus allowing us to determine whether diversifying selection is the dominant force in maintaining polymorphisms in the GPCR US28.
Test for Selective Neutrality
To test whether the frequency spectrum of mutations conformed to the expectations of the standard neutral model, three test statistics were performed: (1) Tajima's D statistic, which considers the difference between estimates based on the number of segregating sites [Watterson, 1975; θw] and the estimates of theta based on the pairwise nucleotide differences among the sequences [Nei, 1987: θπ]; (2) Fu and Li's D* statistic, which was used to compare the observed number of singleton polymorphisms with those expected under a neutral model; and (3) Fu and Li's F* test statistic, which is based on the differences between the number of singletons and the average number of nucleotide differences between pairs of sequences. The DNAsp ver 5 [Librado and Rozas, 2009] software package was used to estimate these summary statistics. Using the same program the sliding window analysis (SWA) was performed with a window length and step size of 50 and 10 sites, respectively.
Positively selected codons were detected using the Fixed-Effect Likelihood (FEL) via the Datamonkey web server [Pond and Frost, 2005] and using the ML approach implemented in CODEML of PAML package version 3.15 [Yang, 1997]. For FEL analysis, P-values <0.05 were used to support positive selection. For PAML analysis, the likelihood ratio test was used to compare M1a, M7, and M8a models that assume no positive selection (ω < 1) with the M2a and M8 models that assume positive selection (ω > 1). Sites with ≥95% Bayes Empirical Bayes (BEB) posterior probabilities are considered as positively selected.
RESULTS
Patterns of Genetic Polymorphisms
A total of 59 unique nucleotide sequences defined by 127 polymorphic sites were recovered from 103 sequences. Of the 127 polymorphic sites, 108 and 19 sites were parsimony informative and singletons, respectively. The general time reversible (GTR) model with gamma shape parameter (G = 0.8289) and invariable sites (I = 0.801) was the best-fit nucleotide substitution model selected by AIC. The estimated substitution rate for A-C, A-G, A-T, C-G, C-T, and G-T are 5.12, 36.9, 2.53, 1.73, 22.84, and 1.0, respectively. The inferred ML tree appeared to be a star phylogeny (not bifurcating with strong bootstrap support) and did not show any evidence of genetic clustering based on geographic affiliation of patients or based on the patient's infection status (single or dual) (Fig. 1).
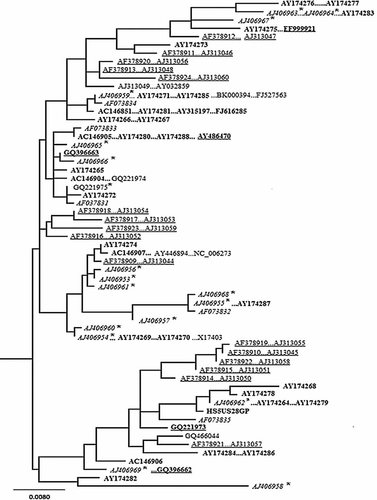
Maximum likelihood tree inferred from complete nucleotide sequence data of the glycoprotein gene, which shows phylogenetic relationships among different isolates of HHV5 sampled from single and dual (with HIV) infected patients from different geographic regions. Patients infected with HIV are marked with an asterisk. Sequences from USA are in bold, from China are underlined, France: Italic; UK: plain text, and from Germany bold with underline.
The mean pairwise nucleotide difference among the sequences is 19.2 ± 8.6. The genetic diversities based on the mean pairwise nucleotide difference (θπ) and based on the number of segregating sites (θw) are 0.0185 ± 0.009 and 0.025 ± 0.006, respectively. Tajima's D, Fu & Li's F and Fu & Li's D* are 0.995, −0.111, and 0.48055, respectively, and are not significantly different from zero (P > 0.05). These findings thus failed to reject the neutral selection hypothesis. The SWA revealed the N-terminal followed by the C-terminal regions showed relatively higher genetic diversities and the middle regions are relatively conserved (Fig. 2a). Consistently, the Tajima's D, although not significant, is positive at the N- and C-terminal regions (Fig. 2b). The positive D value indicates evidence of balancing selection. To know whether any specific codons are under positive selection, ML-based codon-specific selection analyses were performed.
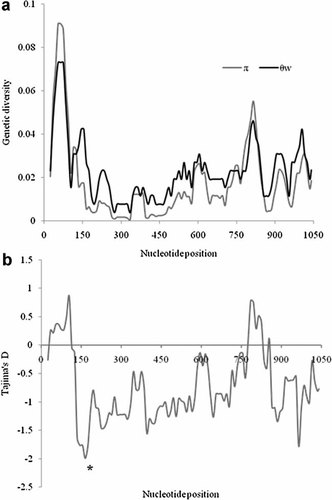
Sliding window analyses (window size = 50, step size = 10) showing (a) the degree of genetic variation estimated based on the number of segregating sites (θw) and the number of mutational differences (π) across the complete glycoprotein gene of US28, and (b) showing the Tajimas' D values for each region. Regions where Tajima's D is significant (P < 0.05) are shown with an asterisk.
Test for Positive Selection
The ML approach implemented in CODEML (PAML package version 3.15) was used to determine whether any of the codons in the US28 region of HCMV have evolved under positive selection. Likelihood ratio tests (LRTs) indicated that positive selection models M2a and M8 are the best-fit models when compared to their corresponding neutral models M1a, M7, and M8a (Table Ia). Three codon sites (18, 19, 190) were inferred to be under positive selection, with BEB posterior probabilities > 99% (Table Ia). Under the M8 model, sites 267 and 329 were also detected as positively selected with BEB posterior probability ≥95%. Consistent with PAML, the FEL method has also detected sites 18, 19, and 190 under positive selection. US28 belongs to the large family of seven-transmembrane spanning G protein-coupled receptors (GPCRs; Fig. 3). The first 34 amino acids constituting the extracellular N-terminal domain confer ligand binding, whereas the last 59 residues forming the intracellular C-terminus regulate receptor expression and desensitization. Of the five positively selected sites, three sites (18, 19, and 267) are located in the extracellular domain of US28 (Fig. 3). Of these three positively selected sites, two sites (E18D/L, D19A/E/G) are located at the N-terminus of the US28 protein and one site (R267K/Q) is located at the third extracellular domain of US28. Only one positively selected site (R329Q/W) is located at the C-terminus. Interestingly, one of the positively selected sites (V190I) is located in the highly conserved transmembrane (TM) domain (Fig. 3).
| Models compareda | −2Δlb | Parameters estimated under selection modelc | Positively selected sitesd |
|---|---|---|---|
| a | |||
| M1a and M2a | 27.05 (df = 2) | p1: 0.92, ω0: 0.03 | 18, 19, 190, 267, 329 |
| P < 0.001 | p2: 0.06, ω1: 1.00 | ||
| p3: 0.02, ω2: 5.42 | |||
| M7 and M8 | 26.30 (df = 2) | p0 = 0.97828, p = 0.08470, q = 0.83795 | 18, 19, 190, 267, 287, 329, 346 |
| P < 0.001 | (p1 = 0.02172), ω = 4.68132 | ||
| M8 and M8a | 24.61 (df = 1) | p0 = 0.97828, p = 0.08470, q = 0.83795 | 18, 19, 190, 267, 287, 329, 346 |
| P < 0.001 | (p1 = 0.02172), ω = 4.68132 | ||
| Codon | dS | dN | dN/dS | Normalized dN − dS | P-Value |
|---|---|---|---|---|---|
| b | |||||
| 18 | 0 | 2.598 | Infinite | 7.621 | 0.0486 |
| 19 | 0 | 4.039 | Infinite | 11.850 | 0.0246 |
| 190 | 0 | 3.007 | Infinite | 8.823 | 0.0193 |
- dS, synonymous substitution rate at the site; dN, non-synonymous substitution rate at the site; normalized; dN − dS; dN − dS divided by the total length of the appropriate tree. P < 0.05 indicates codon under positive selection.
- a Null models (no positive selection): M1a, M7, and M8a, selection models: M2a and M8.
- b P values presented are for rejection of models M1a, M7, and M8a; df = degree of freedom; 2Δl = likelihood score.
- c M2 parameters: numerical values for site categories (p0, p1, and p2) indicate the proportion of codons in the category. Proportion of codons (p0, p1, and p2) having ω (dn/ds) = ω0, ω1, and ω2, respectively. M8 parameters: Proportion of codons (p1) having ω > 1. p and q are parameters that determine the shape of the beta distribution of ω values in models M7 and M8.
- d Posterior probabilities (Bayes Empirical Bayes): >0.99 are in bold, >0.95 and <0.99 are underlined, >0.90 and <0.95 are in plain text.
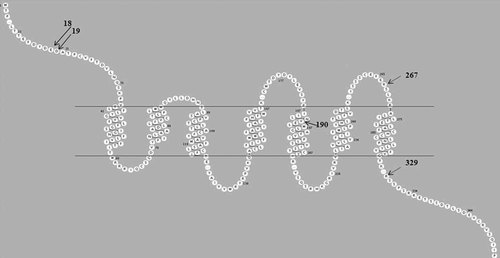
Location of US28 positively selected codon sites in extracellular receptor protein domains. Arrows indicate the location of positively selected sites in US28. Sites 18, 19, and 190 were selected by both FEL and PAML, whereas sites 267 and 329 were selected only by PAML. The seven transmembrane regions are between the two horizontal lines. Sites 18, 19, 267, and 329 are in the extracellular domain, whereas site 190 is located in the transmembrane domain. Diagram created with RbDe online software application [Skrabanek et al., 2003].
DISCUSSION
Like most virally encoded chemokine seven-transmembrane (7TM) receptors, the US28 HCMV encoded 7TM receptor has developed a variety of mechanisms for host immune system evasion, cellular transformation, tissue targeting and possibly for cell entry [Rosenkilde et al., 2008]. In addition, US28 was reported previously to be involved in pathogenic phenotypes such as cardiovascular disease [Muhlestein et al., 2000], HIV entry [Pleskoff et al., 1997], and oncogenic development [Cinatl et al., 2004; Soderberg-Naucler, 2006]. In this context, US28 might have developed genetic polymorphisms as useful devices to evade host immune surveillance [Bodaghi et al., 1998]. In the present study, the pattern of genetic polymorphisms, the extent of geographical associations of clinical isolates, and the underlying genetic mechanisms that maintain diversity in the US28 gene sequence were examined. The results revealed no strong geographic clustering of clinical isolates, which is also the characteristic of UL146 and UL139 [Bradley et al., 2008], as well as UL73 (encoding glycoprotein N) [Pignatelli et al., 2003].
Previous studies provided evidence of genetic polymorphisms in US28 [Arav-Boger et al., 2002; Goffard et al., 2006; Xia et al., 2006]; however, it is unclear as to how these polymorphisms were maintained. The present study revealed that certain codons are under positive selection, and more importantly most of these positively selected sites are located in the extracellular domain of US28. Regions containing codon-18 and -19, located in the extracellular domain of the N-terminus, contributed to the binding of CC- and CXC-chemokines [Casarosa et al., 2003; Stropes and Miller, 2008], as well as the effects of expression and trafficking of US28 [Casarosa et al., 2005]. These regions are thus expected to be under differential selective pressures to maintain conformation and functionality of ligand binding and signaling [Kunstman et al., 2003; Metzger and Thomas, 2010]. The motif mutations in the N-terminus region containing codon 18 and 19 of US28 may be associated with congenital disease [Arav-Boger et al., 2002] or HIV infection [Goffard et al., 2006]. These results suggest that US28 variability can potentially be used as a marker of HCMV infection. US28 variability in a HIV infected population may be due to the highly relevant immunocompromised status of the subjects, which allows the propagation of less virulent cytomegalovirus strains. Thus, it may be that these codons might have played a critical role in evading host immune surveillance; however, further experiments are required to evaluate the putative role of these two codons in other immunocompromised patients (such as organ transplant recipients). The extracellular domains of US28 were exposed to pathogen or chemokine environments, and therefore they would be expected to be involved in host immune system evasion. Point mutations in the third extracellular domain of US28 (K257V, E266V, R267V) abolished its HIV-1 co-receptor activity, while these changes had no apparent effect on the fusion-enhancing activity [Pleskoff et al., 1998].
US28 functional selectivity is attributable to both the specific extracellular chemokine environment and the intracellular complement of G-proteins, resulting in a wide variety of signaling and cell motility responses. In contrast to the N-terminus, only one codon (R329Q/W) in the C-terminus of US28 was under positive selection. Previous studies on US28 have identified the C-terminus as being important for both constitutive receptor phosphorylation and internalization [Miller et al., 2003], as well as US28 pro-migratory signaling [Stropes et al., 2009]. These previous results coupled with the findings presented here suggest that the C-terminus of US28 is especially relevant to studies of the evolution of structure and function of receptors for endogenous signaling.
One of the positively selected sites (V190I) is located at the fifth transmembrane (TM-5) domain of US28. Previous studies [Hubbell et al., 2003; Schwartz et al., 2006] on toggle-switch models of 7TM receptor activation reported that the outer segment of TM-5 may be involved in TM-3, TM-6, and TM-7 toggle switch action during receptor activation. However, the role of TM-5 in toggle switch action is still unclear. The spread of these three helices on the intracellular face of the receptor allows for the binding of signaling proteins such as G-proteins, which promotes subsequent intracellular signal transduction. Therefore, TM-5 may be under strong purifying selection to maintain receptor activation.
In conclusion, the results provide evidence that certain codons in the extracellular domains have evolved adaptively. Such adaptive evolution of those codons could be attributed to the involvement of the extracellular domain in host immune system evasion and ligand-binding activity. In contrast, the intracellular domains are constrained more selectively. These results suggest that differential selective pressures have shaped evolutionary patterns in the functional domains of US28.
Acknowledgements
We thank two anonymous reviewers and Dr. Peggy Hill and Dr. Daniel Elleder for critical reading and thoughtful comments towards improving this manuscript.




