Expression profiles of genes associated with viral entry in HCV-infected human liver†
Study design, sample collection, supervision of laboratory work, data analysis, manuscript writing, and administrative work were done by M. Nakamuta; T. Fujino, R. Yada, Y. Aoyagi, and K. Yasutake performed the laboratory work; M. Kohjima, K. Fukuizumi, T. Yoshimoto, and N. Harada did sample collection; M. Yada, M. Kato, and K. Kotoh did the supervision of laboratory work and data analysis; A. Taketomi, and Y. Maehara did sample collection; M. Nakashima critically reviewed the manuscript; M. Enjoji did study design, data analysis, manuscript writing, and administrative work.
Abstract
Recent studies have demonstrated that several cellular factors are involved in entry of hepatitis C virus (HCV) into host cells. Detailed gene expression profiles of these factors in HCV-infected livers have not been reported for humans. Transcriptional levels of LDL receptor (LDLR), CD81, scavenger receptor class B type I (SR-BI), claudin-1, and occludin genes in liver samples from patients with chronic hepatitis C were investigated. Serum levels of LDL-cholesterol (LDL-C) and HCV core antigen were also evaluated, and expression of claudin-1 and occludin were immunohistochemically analyzed. Compared with normal liver, transcription of LDLR and claudin-1 genes was significantly suppressed (P < 0.0001) and occludin transcription was significantly up-regulated in HCV-infected livers (P < 0.0001). Significant positive correlations were found for LDLR versus occludin, LDLR versus claudin-1, occludin versus claudin-1, and CD81 versus SR-BI in HCV-infected (P = 0.0012, P < 0.0001, P = 0.0004, and P < 0.0001, respectively) and normal livers (P < 0.0001, P = 0.0051, P < 0.0001, and P < 0.0001, respectively). Positive correlation was observed between serum levels of HCV core antigen and LDL-C (P = 0.0147), with their levels negatively correlated to LDLR (P = 0.0270 and P = 0.0021, respectively). Immunohistochemically, hepatocellular expression of claudin-1 and occludin was increased in HCV-infected livers. Different levels of expression were demonstrated at the mRNA and protein levels for occludin and claudin-1 in HCV-infected and normal livers. Correlation of elements associated with viral entry was comparable in HCV-infected and normal livers. J. Med. Virol. 83:921–927, 2011. © 2011 Wiley-Liss, Inc.
Abbreviations used:
LDL-C, LDL-cholesterol; LDLR, LDL receptor; MTP, microsomal triglyceride transfer protein; RBBP6, retinoblastoma binding protein 6; RT-qPCR, quantitative real-time reverse transcription-polymerase chain reaction; SR-BI, scavenger receptor class B type I.
INTRODUCTION
The cellular factors or receptors required for hepatitis C virus (HCV) entry/infection have recently been reported and LDL receptor (LDLR), CD81, scavenger receptor class B type I (SR-BI), claudin-1, and occludin are considered to be essential molecules for HCV entry into hepatocytes [Owen et al., 2009; Perrault and Pécheur, 2009; Pietschmann, 2009; Tang and Grisé, 2009]. A current model predicts a multistep process for entry and infection. (i) The HCV particle consists of a nucleocapsid that is surrounded by a lipid bilayer in which the envelope formed by E1/E2 heterodimers is anchored to a host-cell-derived lipid membrane [Perrault and Pécheur, 2009]. The majority of HCV in the blood is found associated with β-lipoproteins. HCV particles associated with apolipoprotein E non-specifically attach to the cell surface molecules, LDLR, and glycosaminoglycans, an event that precedes virus interaction with CD81 and SR-BI [Burlone and Budkowska, 2009; Owen et al., 2009; Perrault and Pécheur, 2009]. (ii) The next step is specific binding of HCV envelope proteins to the entry receptors CD81 and SR-BI, which localize to the basolateral surface of polarized epithelial cells and interact with HCV envelope protein E2 [Nakagawa et al., 2004; Levy and Shoham, 2005]. CD81/E2 engagement triggers Rho family GTPase-dependent actin rearrangements, which allows lateral movement of the CD81/E2 complex to the tight-junctions, where the CD81/E2 complexes come into contact with claudin-1 and occludin [Brazzoli et al., 2008]. (iii) Claudin-1 and occludin, which are critical components of tight-junctions, have been identified as critical HCV hepatocyte entry factors at a late event, presumably after virus binding and interaction with CD81, and could be crucial factors for HCV internalization [Evans et al., 2007; Benedicto et al., 2009; Liu et al., 2009; Ploss et al., 2009]. No direct HCV–claudin-1 or HCV–occludin interaction has been demonstrated [Evans et al., 2007; Ploss et al., 2009; Krieger et al., 2010], but claudin-1 and occludin co-immunoprecipitate with both HCV envelope proteins, and also form complexes with CD81 and LDLR. Therefore, entry steps mediated by claudin-1 and occludin involve similar mechanisms that take place at the tight junction [Benedicto et al., 2008; Yang et al., 2008]. The tight-junction protein claudin-1 localizes to the basolateral surfaces of hepatocytes [Reynolds et al., 2008] and the non-junctional claudin-1 may be involved in HCV entry [Evans et al., 2007; Cukierman et al., 2009]. Claudin-1 associates with CD81, and this complex is essential for HCV infection [Harris et al., 2008; Harris et al., 2010]. (iv) The final step is presumably clathrin-dependent endocytosis, followed by transport of the viral particles to the endosome [Owen et al., 2009; Perrault and Pécheur, 2009; Pietschmann, 2009; Tang and Grisé, 2009].
Subsequently, non-HCV-permissive human and non-human cell lines become susceptible to HCV when CD81, SR-BI, claudin-1, and occludin are expressed, and CD81 and occludin function as human-specific HCV entry factors [Ploss et al., 2009]. However, the genes for these viral-entry-associated proteins in HCV-infected liver have not been thoroughly investigated in humans. In this study, the transcriptional levels of LDLR, CD81, SR-BI, claudin-1, and occludin genes were examined in liver tissue samples obtained from patients with chronic HCV infection, and their reciprocal relationships were estimated. The expression of claudin-1 and occludin proteins in liver tissue was immunohistochemically analyzed. The level of correlation for these viral entry-associated genes with serum concentrations of LDL-cholesterol (LDL-C) and HCV core antigen were also investigated. LDL-C levels may be related to HCV infection and possibly correlate with the outcomes of interferon treatment [Economou et al., 2008; Torres and Harrison, 2008; Sezaki et al., 2009]. This is the first report investigating the expression profiles of genes associated with HCV entry in HCV-infected human livers.
MATERIALS AND METHODS
Tissue samples were obtained by liver biopsy from 103 patients with chronic hepatitis C prior to any antiviral treatment. These patients were admitted to the Kyushu Medical Center and Kyushu University Hospital in 2007–2009. The background characteristics of these patients are shown in Table I. As a control, normal liver tissue was obtained from 35 living donors that had undergone liver transplantation and whose liver function tests and histological findings were completely normal. The study protocol was approved by the Ethics Committee of Kyushu Medical Center and Kyushu University Hospital, and written informed consent was obtained from all patients.
| Number | 103 |
| Sex (male/female) | 40/63 |
| Age (years) | 56.06 ± 11.65 (23–74) |
| HCV genotype (1b/2a or 2b) | 62/41 |
| Grading (A0/A1/A2/A3) | 0/50/52/1 |
| Staging (F0/F1/F2/F3/F4) | 5/48/25/24/1 |
| Total cholesterol (mg/dl) | 177.37 ± 32.96 (114–265) |
| Triglyceride (mg/dl) | 94.32 ± 38.41 (34–185) |
| HDL-cholesterol (mg/dl) | 54.15 ± 15.45 (25–106) |
| LDL-cholesterol (mg/dl) | 104.14 ± 27.79 (45–167) |
| ALT (IU/l) | 69.76 ± 79.84 (9–482) |
| HCV core antigen (fmol/l) | 7214.37 ± 7724.04 (20–37500) |
Blood samples for serum biochemistry were collected on the day of liver biopsy. Serum levels of HCV core antigen were measured using a chemiluminescent enzyme immunoassay (Lumipulse Ortho HCV Ag; Ortho Clinical Diagnostics, Tokyo, Japan).
Gene expression was examined by quantitative real-time reverse transcription-polymerase chain reaction (RT-qPCR) and compared between HCV-infected and normal livers. The PCR primers for the amplification of LDLR, CD81, SR-BI, claudin-1, occludin, and retinoblastoma-binding protein 6 (RBBP6) are listed in Table II. Total RNA was prepared from the tissue samples using TRIzol reagent (Invitrogen, Carlsbad, CA), and cDNA was synthesized from 1.0 µg RNA using a GeneAmp RNA PCR kit (Applied Biosystems, Branchburg, NJ) in conjunction with random hexamers. The RT-qPCR was performed with a LightCycler-FastStart DNA Master SYBR Green 1 kit (Roche, Basel, Switzerland) in accordance with the manufacturer's instructions. The reaction mixture (20 µl) contained LightCycler-FastStart DNA Master SYBR Green 1, 4 mM MgCl2, 0.5 µM upstream and downstream PCR primers, and 2 µl first-strand cDNA as a template. To control for variations in the reactions, all PCR data were normalized against RBBP6 expression [Chen et al., 2008]. All PCRs were performed in triplicate. The gene expression levels are shown as the relative ratios to those in normal liver and the results are expressed as means ± standard deviation (SD). Continuous variables were compared using the Mann–Whitney U-test. Correlations were evaluated by Spearman's rank correlation coefficient. Values of P < 0.05 were considered statistically significant.
| Genes | Forward (5′→3′) | Reverse (5′→3′) |
|---|---|---|
| LDLR | CAACGGCTCAGACGAGCAAG | AGTCACAGACGAACTGCCGAGA |
| CD81 | AAGCAGTTCTATGACCAGGCCCTAC | TGAGGTGGTCAAAGCAGTCAGTG |
| SR-BI | ATGAAATCTGTCGCAGGCATTG | TGCATCACCTTGGGCATCA |
| Claudin-1 | GCATGAAGTGTATGAAGTGCTTGGA | CGATTCTATTGCCATACCATGCTG |
| Occludin | AAGAGTTGACAGTCCCATGGCATAC | ATCCACAGGCGAAGTTAATGGAAG |
| RBBP6 | GCGACCTGCAGATCACCAA | TGCCATCGCTGGTTCAGTTC |
- LDLR, LDL receptor; SR-BI, scavenger receptor class B type I; RBBP6, retinoblastoma binding protein 6.
Liver biopsy samples from patients with chronic hepatitis C (n = 8) and from living donors for liver transplantation (n = 3) were used for immunohistochemical analysis. Each specimen was fixed in formaldehyde, paraffin-embedded, cut into 5-µm thick sections, and dewaxed in xylene. An endogenous peroxidase block was performed with 3% hydrogen peroxide in methanol for 15 min. For antigen retrieval, tissue slides were incubated with Antigen Retrieval Reagent-Basic (R&D Systems, Minneapolis, MN) at 95°C for 5 min. After cooling to room temperature, monoclonal anti-human claudin-1 (final concentration 1.5 µg/ml; Abcam, Tokyo, Japan) and polyclonal anti-human occludin (final concentration 2 µg/ml; Abcam) were used as primary antibodies. Histofine Simple Stain PO (Nichirei, Tokyo, Japan) was used in our assay in accordance with the manufacturer's protocol.
RESULTS
The transcriptional levels of genes related to entry of HCV in the host cell were examined by RT-qPCR in liver samples and compared among 103 patients with chronic hepatitis C (Table I) and 35 healthy individuals. As shown in Figure 1, the transcription of LDLR was markedly suppressed in HCV-infected livers when compared with normal livers (P < 0.0001). Expression levels for genes encoding CD81 and SR-BI were comparable between HCV-infected and normal liver samples. Transcription of the claudin-1 was significantly decreased (P < 0.0001) and for occludin it was clearly increased in HCV-infected liver samples (P < 0.0001). Correlation of the levels of transcription for pairs of genes in HCV-infected and normal livers were evaluated. All pairs with significant correlation are listed in Table III. A strong positive correlation was found between each pair of LDLR, claudin-1 and occludin, and between CD81 and SR-BI, in HCV-infected liver as well as normal liver (Table III, Fig. 2). Serum levels of LCL-C and HCV core antigen showed a significant positive correlation (Fig. 3A). Both serum parameters were negatively correlated with the transcriptional levels of LDLR (Fig. 3B,C), but not with those of other genes examined (data not shown). Formaldehyde-fixed liver samples were incubated with appropriate specific antibodies to ascertain protein expression levels of claudin-1 and occludin. In HCV-infected livers, claudin-1 and occludin were detected and localized to hepatocyte membranes, exhibiting low-level staining. Their expression was not evident in normal liver samples (Fig. 4).
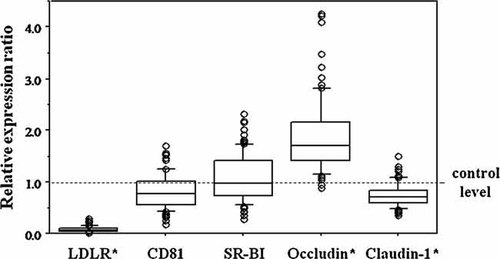
Transcription levels of genes associated with viral entry in HCV-infected livers. The broken line indicates the mean level in normal control subjects. LDLR, LDL receptor; SR-BI, scavenger receptor class B type I. *Significantly different to normal liver (P < 0.01).
| Correlation coefficient | P-value | |
|---|---|---|
| HCV-infected liver (n = 103) | ||
| CD81 versus SR-BI | 0.652 | <0.0001 |
| LDLR versus Claudin-1 | 0.381 | <0.0001 |
| Claudin-1 versus Occludin | 0.341 | 0.0004 |
| LDLR versus Occludin | 0.315 | 0.0012 |
| LDLR versus SR-BI | 0.284 | 0.0036 |
| Normal liver (n = 35) | ||
| LDLR versus Occludin | 0.724 | <0.0001 |
| Claudin-1 versus Occludin | 0.628 | <0.0001 |
| CD81 versus SR-BI | 0.620 | <0.0001 |
| LDLR versus Claudin-1 | 0.463 | 0.0051 |
| SR-BI versus Occludin | 0.353 | 0.0372 |
| SR-BI versus Claudin-1 | 0.337 | 0.0463 |
- LDLR, LDL receptor; SR-BI, scavenger receptor class B type I.
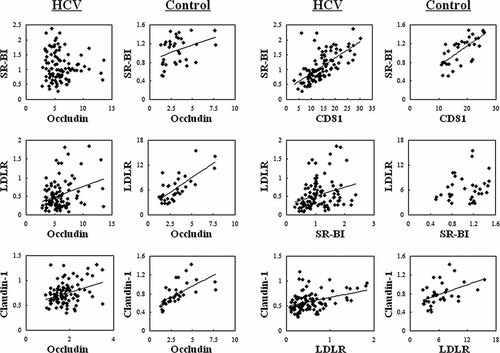
Correlation analysis of gene expression levels in HCV-infected (HCV) and normal (control) livers. SR-BI, scavenger receptor class B type I; LDLR, LDL receptor.
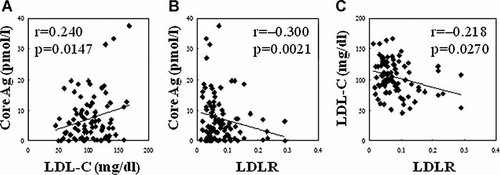
Correlation of serum levels for LDL-cholesterol (LDL-C) versus HCV core antigen (Core Ag, A), gene expression levels of LDL receptor (LDLR) versus serum levels of Core Ag (B), and gene expression levels of LDLR versus serum levels of LDL-C (C).
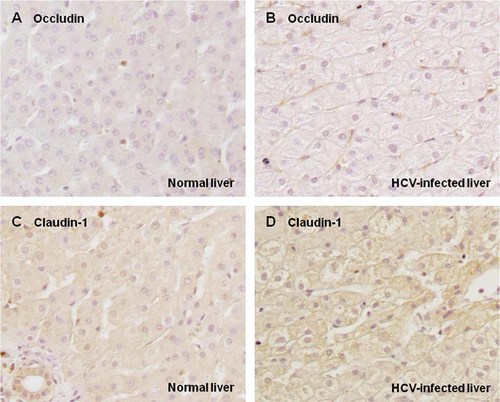
Immunohistochemical staining of normal (A,C) and HCV-infected liver tissue (B,D) for occludin (A,B) and claudin-1 (C,D). ×200 magnification (A,B), ×100 magnification (C,D).
The levels of core antigen varied with genotypes (genotype 1, 7715.84 ± 7774.92 fmol/l; genotype 2, 5917.54 ± 7145.22 fmol/l) but the results described above were independent of HCV genotype or viral quantity (data not shown). Moreover, the results were independent of sex, histological grading, or staging (data not shown).
DISCUSSION
To the best of our knowledge, there have been no human studies regarding the effect of HCV infection on the expression profile of genes associated with viral entry in the liver. Here, we present the results from patients with chronic hepatitis C. Our data were not simply from hepatocytes, because the biopsied liver samples contained other cell types and HCV RNA is often found in lymphocytes [Durand et al., 2010; Stamataki, 2010]. However, hepatocytes were in the majority for the samples and the obtained data were independent of the grade of lymphocytic infiltration (grading). Therefore, it was considered that the data were representative of those from hepatocytes.
The transcription levels of LDLR were markedly suppressed in HCV-infected livers and were inversely correlated with the serum levels of LDL-C and HCV core protein (Figs. 1 and 3). The levels of LDL-C and HCV core protein were positively correlated (Fig. 3). It has been demonstrated that HCV inhibits the activity of microsomal triglyceride transfer protein (MTP), which results in down-regulation of VLDL secretion, hypobetalipoproteinemia and hypocholesterolemia [Perlemuter et al., 2002; Huang et al., 2007; Syed et al., 2010]. Because HCV virion assembly and secretion utilize the VLDL pathway, and HCV particles are found in the blood as lipoprotein complexes [Huang et al., 2007; Syed et al., 2010], positive correlation between serum levels of HCV core antigen and LDL-C in this study was compatible with previous observations. Suppressed expression of LDLR in HCV-infected livers may reflect intracellular overload of cholesterol and triglyceride, which arises from reduced MTP activity and increased lipogenesis in hepatocytes [Brown and Goldstein, 1981; Goldstein and Brown, 1990; Nakamuta et al., 2009], and it has been observed that expression of LDLR is inversely related to the concentration of LDL-C [Goldstein and Brown, 2009]. Accordingly, the presented results regarding the levels of LDLR, LDL-C, and HCV core antigen confirm the expression profiles described in previous studies.
The expression of other HCV receptors involved in cell entry was also examined. In vitro studies have shown conflicting results in the expression profile of these receptors. For example, one group has reported slightly up-regulated claudin-1 expression [Reynolds et al., 2008] and another has shown down-regulated expression of claudin-1 and occludin [Liu et al., 2009] in HCV-infected Huh7.5 cells. The use of cultured cells as a model for infection does not fully represent the situation in the human liver because polarity is crucial to liver function, and Huh7 cells are relatively nonpolarized [Brazzoli et al., 2008]. Reynolds et al. [2008] have reported the immunohistochemical expression of CD81, SR-BI, and claudin-1 in human liver, with no significant changes noted in CD81 and SR-BI expression levels between normal and HCV-infected livers. However, hepatocellular levels of claudin-1 were increased in HCV-infected livers compared with normal livers [Reynolds et al., 2008]. In this study, no significant difference was observed in the expression of CD81 and SR-BI between normal and HCV-infected livers at the transcriptional level (Fig. 1). In terms of tight-junction proteins, the transcriptional levels of the claudin-1 gene were down-regulated and those of the occludin gene were up-regulated in HCV-infected livers as compared to normal livers (Fig. 1). Immunohistochemical staining revealed that expression of claudin-1 and occludin appeared to increase in hepatocyte membranes of HCV-infected livers (Fig. 4). In general, antigen retrieval is required when attempting immunohistochemical detection of claudin-1 and occludin; therefore, expression levels should be carefully evaluated. The most suitable method for antigen staining is yet to be established and a greater number of samples need to be analyzed; however, the immunohistochemistry results presented in this study may be reliable because claudin-1 staining was similar to that demonstrated by Reynolds et al. [2008]. The staining levels appeared to be able to discriminate between HCV-infected and normal livers. In this study, there was a discrepancy between mRNA and protein levels for claudin-1. While the reason for this cannot yet be definitively explained, previous studies have suggested that HCV infection could directly increase translation of claudin-1 without altering mRNA levels, and that claudin-1 expression and localization might be regulated at the level of protein phosphorylation and palmitoylation [Yamauchi et al., 2004; Van Itallie et al., 2005; Reynolds et al., 2008].
Correlation analysis of the transcriptional levels of viral entry-associated genes demonstrated significant positive correlation for LDLR versus occludin, LDLR versus claudin-1, occludin versus claudin-1, and CD81 versus SR-BI in HCV-infected and normal livers (Table III, Fig. 2). In particular, the relationship between occludin and claudin-1 (tight-junction proteins) and between CD81 and SR-BI (E2-binding receptors) could reflect their functional similarity as viral receptors. Moreover, these findings indicate that the reciprocal relationship and regulatory pathways of genes associated with viral entry in the normal liver are consistent even during HCV infection. Only the correlation between LDLR and SR-BI appeared to be induced by HCV infection (Table III, Fig. 2), although any reasons cannot be defined. Marked suppression of LDLR following HCV infection may be associated with SR-BI expression. In any case, further studies are required to elucidate the interplay between HCV and the known host factors.
Expression profiles of genes associated with HCV entry were investigated in HCV-infected and normal livers in humans. Transcription of LDLR and claudin-1 genes was significantly suppressed and that of occludin was significantly up-regulated in HCV-infected livers. The LDLR levels were inversely correlated with the serum levels of LDL-C and HCV core protein in HCV-infected livers. Correlation of elements associated with viral entry was comparable in HCV-infected and normal livers.




