Human papillomavirus 16 variants from Zambian women with normal pap smears
Abstract
Human papillomavirus (HPV) type 16 is the most prevalent high-risk viral genotype associated with cervical cancer. Six distinct phylogenetic clusters of HPVs have been identified and are distributed differently across five continents. HPV16 DNA was extracted from cervico-lavage samples from women with normal pap smears. The LCR regions were amplified in triplicate, cloned, sequenced, and analyzed from a total of 11 recovered HPV16 positive samples [Ng'andwe et al. (2007): BMC Infect Dis 7:77] were analyzed for sequence variation. The HPV16 LCR variants were assessed for promoter activity by use of a luciferase reporter gene. Six novel HPV16 variants with nucleotide exchanges in the LCR region were identified. Five clones were classified as European group HPV16 variants and one as an African group variant. Two of these variants had relatively lower promoter activity, 30% of that of the wild-type strain. The decreased promoter activity of some HPV16 variants may decrease expression of viral oncogenes and may be linked with the development, phenotype and severity of the cervical lesions in women infected with these across HPV16 variants. J. Med. Virol. 83:1230–1237, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Epidemiological and molecular studies have demonstrated that high-risk alpha-human papillomavirus (HPV) is the etiological agent responsible for the majority of cervical cancers, the second most common female cancer globally [Walboomers et al., 1999; zur Hausen, 2000; Munoz et al., 2003]. So far, more than 120 different HPV types have been identified and characterized on the basis of DNA sequence analysis [de Villiers et al., 2004]. At least 40 HPV types infect the genital area, of which 15 types (HPV16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -59, -66, -68, and -73) are classified as high-risk because of their oncogenic potential [Munoz et al., 2003]. HPV16 is the most prevalent high-risk type associated strongly with cervical cancer and accounts for about 50–60% of all cases of cervical cancer globally [Clifford et al., 2003]. HPV16 has also been found in multiple cancers of the anus, vulva, vagina, and penis, as well as a subset of head and neck carcinomas [Koskinen et al., 2003; Ritchie et al., 2003].
Although HPV genomes have been thought to be rather evolutionarily stable, nucleotide changes have been increasingly observed in clinical isolates. An HPV variant is defined as one that differs from other viruses of the same type by up to 2% in conserved regions of the genome, such as E1 or L1 ORFs, and by up to 5% in the LCR [Kammer et al., 2000]. HPV16 has been examined in great detail due to its medical importance, and numerous HPV16 variants have been identified. Studies and phylogenetic characterization of the LCR of HPV16 from worldwide cervical samples demonstrated that HPV16 variants can be divided into six distinct phylogenetic clusters distributed differently across the five continents: the European (E) group, two African (Af) groups, the Asian (As), Asian American (AA), and North American (NA) variants [Wheeler et al., 1997; Yamada et al., 1997]. Different HPV16 variants exhibit differences in their biological and biochemical properties. Epidemiologic studies indicate that non-European variants of HPV16 exhibit increased oncogenicity, specifically members of the Asian-American class AA [Kammer et al., 2000].
The early proteins of high-risk HPV, E6 and E7, play an important role in the carcinogenesis of human cells. They bind with and inhibit the function of the tumor suppressors p53 and pRb respectively [Dyson et al., 1989; Scheffner et al., 1990]. The high-risk E6 and E7 protein also modify the expression activity of many cellular proteins in order to promote cell proliferation [Underbrink et al., 2008]. The expression level of HPV E7 is closely correlated with the transforming potential of the virus, and high levels of HPV E7 mRNA expression have been found in oral and cervical cancer biopsy specimens [Liu et al., 1995]. Recently, studies have shown that HPV16 E5 also plays a critical role in mechanism of tumorigenesis [Hu et al., 2009]. The E1 and E2 proteins of HPV are required for both replication of viral DNA and transcriptional regulation [Longworth and Laimins, 2004]. The Long control region (LCR), an 800 bp long non-encoding region of HPV genome, contains an epithelial cell-specific enhancer, putative binding sites for cellular and viral transcription factors and the P97 promoter at its E6-proximal end. The transcription of HPV16 early genes initiates at the p97 early promoter and is controlled by a complex interaction of many cellular and viral factors that bind to the regulatory region in the LCR. Some of these factors include; YY1, NF1, SP1, AP1, TEF1, Oct-1, GRE, and NF-IL6 [Chan et al., 1990; Chong et al., 1991; Ishiji et al., 1992; Apt et al., 1994; May et al., 1994; O'Connor and Bernard, 1995; Khare et al., 1997]. P97 activity plays the central role in the pathogenesis of HPV16. Exchange of nucleotides within transcription factor binding sites resulting in an increase in viral promoter activity and enhanced viral tumorigenesis, has been previously reported for HPV16 [Dong et al., 1994; Kammer et al., 2000; Tornesello et al., 2000]. One study reported that altered p97 activities are also primarily responsible for the strong differences in replication levels in HPV16 LCR variants [Hubert, 2005].
Although almost all cases of cervical cancer are attributable to HPV infection, infection alone is not sufficient to cause cancer. HPV DNA has often been found in normal cervical tissue [Johnson et al., 1990; Woods et al., 1993]. The rate of HPV infection and cervical cancer is highest in the African continent. A previous study investigated the distribution of HPV genotypes in a population of Zambians with normal pap smears [Ng'andwe et al., 2007]. That study showed that HPV16 and HPV18 were the two most prevalent types, and each was present in 21.6% of Zambian samples, significantly higher than the average world rate [Ng'andwe et al., 2007]. In the present study, six novel HPV16 variants were identified by sequence changes in the LCR. HPV variants were isolated from vaginal lavage specimens from Zambian women with normal pap smears. To correlate LCR mutations with changes in p97 promoter activity, a luciferase reporter gene was cloned under the control of each LCR variant or the LCR of the HPV16 W12E isolate. The W12E HPV16 isolate falls within the European group and is widely used as a prototype for HPV16 studies since the full-length genome was cloned [Flores et al., 1999]. LCR variants that contained multiple mutations were systematically analyzed for the contribution of each point mutation to LCR promoter activity.
MATERIALS AND METHODS
Study Participants
This study reports an analysis of HPV16 variant distribution among HIV positive and negative women, sampled from a previously established cohort study [Ng'andwe et al., 2007]. All Human subjects protocols were approved by safety committees at the University of Zambia and UNL in accordance with the Helsinki Declaration. Patient participation was entirely voluntary and written consent was required for inclusion in the study. Demographic details of the cohort participants from the metropolitan area of Lusaka were collected and disease histories as well as physical examinations were carried out to rule out any clinical symptoms or visible signs for these conditions. Pap smears were examined and classified according to the pap classification protocol; pap I (normal), pap II (inflammation), pap III (dysplasia), pap IV (carcinoma in situ), and Pap V (carcinoma), as described previously [Ng'andwe et al., 2007]. All of the participants chosen for this study had normal pap smears.
Sample Collection
Vaginal lavage samples and pap smears were collected from all patients. Vaginal lavage specimens, treated with DNeasy lysis buffer (Qiagen, Valencia, CA) were stored at −20°C. All specimens were then shipped to the Nebraska Center for Virology at the University of Nebraska-Lincoln (UNL) for testing.
DNA Isolation
Total DNA was extracted from vaginal lavage samples using the DNeasy Tissue extraction kit (Qiagen). HPV16 infection in these samples was confirmed by PCR using GP5+/GP6+ and CPI/CPII primers followed by sequencing as detailed in the previous study [Ng'andwe et al., 2007].
PCR Amplification of the LCR
The LCR of HPV16 was amplified from the DNA samples by use of one pair of primers: P-LCR1: 5′-CCGAGCTCACGCAAAAAACGTAAGCTG-3′ (nt 7,133–7,150) with the SacI site underlined and P-LCR2: 5′-CCAAGCTTTCCTAGAACATTGCAGTTCTC-3′ (nt 96–114) with the HindIII site underlined. These primers amplified a DNA fragment of 886 bp, corresponding to the complete LCR sequence of HPV16 (nt 7,133–114). The PCR amplification was performed in 50 µl of the reaction mixture containing 2 µl template DNA, 1.5 mM MgCl2, 10 mM dNTP, 10 pmol of each primer and 5 U of Taq polymerase (Invitrogen, Diego, CA) at the cycle condition of denaturing at 94°C for 50 sec, annealing at 58°C for 50 sec, extension at 72°C for 1 min, totally 30 cycles. PCR products were analyzed on 2% agarose gel and purified with QIAquick Gel Extraction Kit (Qiagen), and then sequenced on both strands for the entire LCR of HPV16. To ensure that identified LCR sequence variants were not the result of Taq-derived mis-incorporations, three independent PCR amplifications, cloning, and sequencing steps were performed. The sequence data was compared with the corresponding fragment of HPV16 W12E isolate by use of BLAST alignment software online (http://blast.ncbi.nlm.nih.gov/).
Plasmid Constructions
The luciferase reporter vector under control of the LCR from HPV16 isolates (pGL3-LCR) were constructed by cloning the entire LCR from HPV16 variants, as well as the W12E isolate into the SacI and HindIII multiple cloning site of pGL3-basic plasmid (Promega, Madison, WI). The generated recombinant plasmids were: pGL3-LCR-101, pGL3-LCR-110, pGL3-LCR-902, pGL3-LCR-374, pGL3-LCR-536, pGL3-LCR-434, and pGL3-LCR-W12E. The recombinant plasmids to be used in transfection experiments were prepared using QIAfilter Plasmid Maxi Kit (Qiagen).
The mutant constructs containing the individual point mutations that existed in the LCR of 374 and 110 HPV16 variants were created using the Quick Change II XL Site-Directed Mutagenesis kit (Stratagene, San Diego, CA). The point mutations were introduced to LCR of HPV16 W12E isolate by PCR with various primers containing mutation at nt 7,405, nt 7,416, nt 7,632, nt 7,742, nt 7,797 in variant 374, and nt 47, nt 7,405, nt 7,826 in variant 110. The parent plasmid was removed by digestion with DpnI, and further transformation was carried out according to the manufacture's instructions. The mutant recombinant plasmids pGL3-374-7405 Mu, pGL3-374-7416 Mu, pGL3-374-7632 Mu, pGL3-374-7742 Mu, pGL3-374-7797 Mu, pGL3-110-47 Mu, pGL3-110-7826 Mu, and pGL3-374-7405 Mu were generated.
Cell Culture and Transient Transfection
HeLa cells and HaCat cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Cells were seeded at a density of 2 × 105 cells per well of six-well plates 1 day prior to transfection. Two micrograms plasmids of pGL3-LCR containing the distinct LCRs from the different HPV16 variants, mutated LCRs or W12E LCR prototype were transfected into monolayer cells together with 1 µg pCMV-β-galactosidase expression plasmid that served as an internal control to normalize for transfection efficiency. The luciferase activity was measured at 48 hr post-transfection with the Luciferase Assay system (Promega) according to the manufacturer's protocol. The relative light units were determined using a luminometer. The expression of β-galactosidase activity was determined using O-nitrophenyl-β-D-galactopyranoside (ONPG) as a colorimetric substrate. Relative luciferase activity measurements of the different plasmids were normalized against β-galactosidase activity. The averages were based on the mean of five independent experiments.
RESULTS
Identification of HPV16 Variants
A total of 11 HPV16 positive patient samples were recovered in a previous study [Ng'andwe et al., 2007]. DNA fragments containing the HPV16 LCR (nt 7,133–114) were amplified from HPV16 positive vaginal lavage specimens of Zambian women who had normal pap smears. We searched for genetic variation in the LCRs from the 11 HPV16 positive samples by DNA sequencing. Five wild-type (European) HPV16 LCRs and six novel HPV16 variants were recovered with nucleotide changes in the LCR sequence, not previously described (Table I). Five new variants; 902, 110, 374, 434, and 536 were classified within the European group and they exhibited 2, 4, 6, 1 and 3 intra-type variations, respectively, compared to HPV16 W12E isolates. The rate of mutation that accounts for 902, 110, 374, 434, and 536 variants in LCR appears to be above the intrinsic cellular mutation rate. Only one variant, 101, classified in the African group, differed by two nucleotides, at nt 7,416 and nt 7,797, from the sequence of the Af1a B2 isolate published by Kammer et al. [2000]. The G to A transition at nt 7,797 was found in all variants except for variant 902. In contrast to nucleotide substitutions in the LCR of HPV variants described previously, several nucleotide insertions were found within LCR sequence of new HPV16 variants identified in this study. These data suggest that the diversity of HPV variants in natural infections has been under-appreciated.
| Position (nt) | W12E | Variants | BS | |||||
|---|---|---|---|---|---|---|---|---|
| 902 | 101 | 110 | 374 | 434 | 536 | |||
| 31 | C | T | Sp1 | |||||
| 47 | (In) A | (In) A | E2 | |||||
| 83 | A | C | ||||||
| 7,405 | (Del) T | (In) C | ||||||
| 7,416 | A | T | (In) G | |||||
| 7,487 | G | A | ||||||
| 7,632 | (In) A | AP1 | ||||||
| 7,687 | C | A | ||||||
| 7,712 | T | A | NF1 | |||||
| 7,742 | T | A | (In) G | |||||
| 7,743 | (In) C | NF-1 | ||||||
| 7,744 | (In) A | |||||||
| 7,762 | C | T | ||||||
| 7,784 | C | T | ||||||
| 7,797 | G | A | A | A | A | A | TEF-1 | |
| 7,826 | (In) A | YY-1, TEF1 | ||||||
| 7,832 | G | T | (In) A | AP1 | ||||
| 7,874 | C | A | ||||||
- Nucleotide changes are indicated within each variant. Insertions are indicated by (In) and deletions by (Del). Predicted transcription factor binding sites “BS” are shown in the right-most column.
Two HPV16 Variants Isolated From Zambian Women Have Relatively Lower Promoter Activities
In order to assess the influence of nucleotide alterations in LCR on promoter activity, we utilized the recombinant luciferase reporter plasmids containing distinct LCR sequences from six isolated HPV16 variants, as well as the HPV16 W12E isolate, each cloned upstream of a luciferase reporter gene (pGL3). Constructs were transiently transfected into HeLa and HaCat cells. The luciferase assay revealed that there were no significant differences in promoter activity of LCRs from 902, 101, 434, and 536 variants compared to that of HPV16 W12E prototype isolate. However, the expression of luciferase under control of LCR from variants 110 and 374 decreased to 30% and 26.6% of that from W12E isolate in HeLa cells, and decreased to 27% and 34% in HaCat cells (Fig. 1). These data suggest that variation in LCR-p97 promoter function are tolerable and potentially important for HPV fitness. Previous studies have found that mutations in the LCR that alter p97 promoter function often significantly affect gene expression as well as viral copy number [Hubert, 2005].
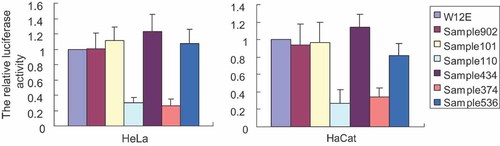
The promoter activity under control of LCR from different variants as well as the W12E isolate is shown. HeLa cells and HaCat cells were transfected with 2 µg pGL3-LCR plasmids containing LCR of HPV16 W12E or different variants. The relative luciferase activity was averaged from five independent experiments in HeLa, HaCat cells and presented relative to that of W12E pGL3-LCR. The data are represented as the mean ± SD, indicated by error bars.
The Contribution of Different Point Mutations in LCR of HPV16 Variants 374 and 110 on Decreased Promoter Activities
To determine which of these point mutations in the LCR of HPV16 variants 374 and 110 decreased promoter activity of the virus, we introduced each different point mutation independently in LCR using the backbone of pGL3-LCR-W12E plasmid. The mutated plasmids were transfected into HeLa cells for analysis. The results showed that in the variant 374, the expression of luciferase under control of the mutated LCR with mutations at nt 7,405, nt 7,416, nt 7,632, and nt 7,797 were little compared to the plasmid with LCR from the W12E isolate. The plasmid containing the mutation at nt 7,742 was responsible for a 70% reduction in the expression of luciferase (Fig. 2), corresponding to the expression level of luciferase under the control of LCR from variant 374 with all five point mutations. These results show that decreased promoter activity of variant 374 was primarily attributed to the point mutation at nt 7,742. In Eletrophoretic mobility shift assays (EMSAs), no differences were found in the binding of nuclear proteins from HeLa cells to DNA probes (from nt 7,733 to nt 7,750) from HPV16 W12E or variant 374 (data not shown), yet minute changes could still be affecting transcriptional activation. In variant 110, the expression of luciferase under the control of the LCR with mutations at nt 7,405 was decreased by about 87.5% and a mutation at nt 7,826 was decreased to about 45%. The decrease in promoter activity in variant 110 may be due to a synergistic effect between the two point mutations that exist in it. The mutation at nt 7,405 and nt 7,826 also did not induce an obvious difference in the binding affinity between nuclear proteins and DNA in the EMSA results (data not shown).
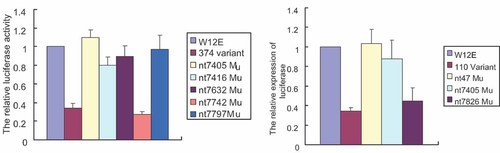
The contribution of different point mutations found within the LCR of 374 or 110 variants on promoter activity compared to the W12E isolate. HeLa cells were transfected with 2 µg pGL3-LCR plasmids containing the LCR of HPV16 W12E or each combination of different point mutations from 374 or 110 variants respectively. The relative luciferase activity is the average of five independent experiments in HeLa cells and presented relative to that of W12E pGL3-LCR. Data are represented as the mean ± SD, indicated by the error bars.
DISCUSSION
In this study, we identified novel HPV16 LCR sequence variants recovered from cervico-vaginal lavage specimens isolated from Zambian women who had no visible evidence of HPV infection. Analysis of the LCR nucleotide sequences showed that five of six HPV16 variants were classified within the European group, while the other one, variant 101, was classified within the African group. Most studies on HPV16 variants have focused on isolates from clinical patients who already had cervical cancer or cervical squamous intraepithelial lesions (SIL). In contrast, the present study brings evidence of a significant number of LCR variant strains of HPV16 actively replicating in asymptomatic women.
We focused on variants of the HPV16 LCR in this study since the p97 promoter has a crucial role in regulating transcription and replication of the virus. Although the direct biological consequence of variation in the LCR of HPV genomes on cervical carcinogenesis is not fully understood, several studies indicate that variants of HPV16 are associated with higher-risk of cervical neoplasias of various stages [Smits et al., 1994; Londesborough et al., 1996; Tornesello et al., 2000; Hildesheim et al., 2001; Burk et al., 2003; Hubert, 2005; Schlecht et al., 2005; Pande et al., 2008]. The mutations accrued in HPV genomes isolated from tumors are diverse in location and type, and thus, the functional outcome is often difficult to predict. However, our current evidence of multiple mutations in the LCRs of actively replicating HPV16 genomes from women with normal pap-smears suggests that these differences are relevant to viral promoter activity and fitness. Relatively few studies have been done on HPV variants from women with normal pap-smears [Schmidt et al., 2001].
Non-European variants, particularly the Asian-American variants, tend to persist more frequently than E variants. In addition, non-E variants were more strongly associated with both the prevalence and incidence of high-grade cervical intraepithelial neoplasia (CIN) lesions and cervical cancer than the E variants [Villa et al., 2000]. Kammer et al. [2000] reported that HPV16 Af1a and 2a variants exhibit p97 promoter activity comparable to the European reference clone. Thus, it may not be that surprising that HPV E and Af1a variants are found in women with normal Pap smears. It is possible that the distribution of variants is not only influencing geographical distribution, but also by severity of the clinical phenotype.
The results showed that six HPV16 LCR variants exhibited differing p97 promoter activity when linked upstream of the luciferase reporter gene. The 374 and 110 variants showed approximately a 70% decrease in p97 promoter activity, whereas other E group variants in this study and Af1a group, 101 variant were more or less as active as the HPV16 W12E promoter. This kind of large decrease in p97 promoter activity observed in variants 374 and 110, although rarely reported, might favor low-copy and long-term persistence of the virus, and reduced probability of cervical cancer.
The importance of several transcription factor binding sites within LCR for transcription and expression of HPV16 early genes has been confirmed in previous studies [Hubert, 2005]. Single nucleotide changes can increase or decrease transcription factor affinity for binding sites or change their ability to interact with accessory proteins. Schmidt et al. [2001] reported that HPV16 LCR variants isolated from cancer cells contain nucleotide changes predominantly within or close to YY1 binding site. Variants obtained from asymptomatic carriers contained different single nucleotide changes, mainly within or close to binding sites of transcription factors such as AP-1, Oct-1, NF-1, TEF-1, TEF-2, SP1, viral E2, and also YY1 [Schmidt et al., 2001]. Not surprisingly, the presented results show that some of the nucleotide exchanges identified in the LCR from the six different HPV16 variants were located near or within putative binding sites for cellular or viral factors. The binding sites in the LCR variants 374 and 110 that resulted in a significant decrease in p97 promoter activity included AP1, NF-1, YY-1, TEF-1, and E2 binding sites (Fig. 3). By systematic analysis of point mutations, we identified the important nucleotide changes in variants 374 and 110 by their effects on p97 promoter activity. Only the LCR clone with nucleotide exchanges in the NF-1 binding site, a G insert at nt 7,742 and an A insert at nt 7,744, led to a decrease in p97 promoter activity to a level corresponding to that of 374 variant. These results indicate that the decreased p97 promoter activity of variant 374 was mainly due to affects on the NF-1 binding site near nt 7,742. As a ubiquitously expressed transcription factor, NF-1 binds to the regulatory elements within the LCR and plays a key role in the transcriptional stimulation of E6/E7 expression through the p97 promoter in HPV [Chong et al., 1991; Apt et al., 1994]. The nucleotide changes at NF-1 binding site that exist in the LCR of variant 374 probably reduce the binding affinity between NF-1 and DNA sequence, thus partly decreasing the transcriptional activity through NF-1. However, the EMSA with HeLa cell nuclear extract was not able to detect differences in protein binding to oligonucleotides with or without mutations at nt 7,742 (as in variant 374 (data not shown)).
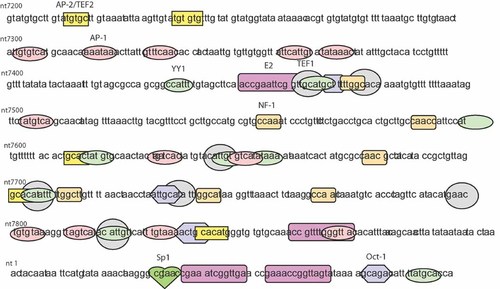
Map of the putative transcription factor binding sites within LCR of HPV16. Each of the transcription factor binding sites are indicated; AP-2, AP-1, YY1, HPV16 E2, TEF1, NF-1, Sp1, and Oct-1.
In variant 110, a point mutation at nt 7,826 (corresponding to YY1 and TEF-1 binding sites) led to a major decrease in promoter activity. YY1 is considered a repressor for HPV16 transcription activity, and several YY1 binding sites have been reported in the HPV16 LCR. Single nucleotide mutations in YY1-binding sites that increase p97 activity three- to sixfold have been identified in HPV16 DNA from cervical carcinomas [Dong and Pfister, 1999]. In this study, we found a mutation at nt 7,826, within the YY1 binding site, which decreased p97 activity by about 50%. The YY1 binding site at nt 7,826 overlaps with a TEF-1 binding site. This may indicate competition for binding, displacement, or steric hindrance between the two proteins.
We found it intriguing that there were a significant number of HPV16 variants circulating in naturally occurring infections. The present results suggest that HPVs may exist in greater evolutionary diversity than previously appreciated. APOBEC3 cytosine deaminases have been described for their role in restricting infectivity of retroviruses by editing the single stranded DNA. Interestingly, a previous study has also shown that these deaminases are able to edit HPV16 DNA [Vartanian et al., 2008]. Furthermore, APOBEC3 expression is known to be induced in the more terminally differentiated keratinocyte layers, where HPV genome amplification takes place. One of the variants we isolated, 101 did not show decreased promoter activity, but had point mutations typical of those induced by APOBEC 3a, 3c, and 3H [Vartanian et al., 2008]. This variant fell within the Africa group, which is thought to be less oncogenic compared to the European group. It is possible that at least some of this variation in HPV genomes is induced by mechanisms such as that described for APOBEC3 cytosine deaminases, but other mechanisms, such as polymerase driven insertions and deletions clearly also impact the pool of variants.
Acknowledgements
This study was supported in part by PHS grant (D43 TW001492), NCI (CA75903) to CW and NCRR COBRE (RR15635) to CW and PA; LY was a Fogarty Scholar.




