Molecular epidemiology of chronic hepatitis B virus infection in Greece
Abstract
Virological data on chronic hepatitis B virus (HBV) infection in Greece are limited. HBV genotypes, surface antigen (HBsAg) subtypes, and HBsAg “a” determinant mutations among patients infected chronically with HBV, were investigated. Serum samples from 135 HBsAg positive patients were tested. Serologic (HBsAg, anti-HBs, HBeAg, and anti-HBe), virologic (HBV-DNA quantitation) and biochemical markers (serum alanine aminotransferase/ALT and aspartate aminotransferase/AST) were analyzed. HBV genotypes and HBsAg subtypes were determined by partial sequencing of the S gene. Genotyping was performed by using the National Center for Biotechnology Information online Genotyping tool and phylogenetic analysis. Nucleotide sequences were aligned pair wise with ClustalW and phylogenetic trees were constructed by the neighbor-joining method. Sequences were also used to predict HBV HBsAg subtypes. In six patients (4%), simultaneous presence of HBsAg and anti-HBs was determined, whereas 47 patients (35%) were HBeAg positive, 84 (62.5%) were anti-HBe positive, and four patients (3%) were characterized by the simultaneous presence of HBeAg and anti-HBe. Mean ALT was 238 IU/L (standard deviation = 576.84), and HBV-DNA levels ranged from 1.02 × 105 to 2.2 × 107 IU/ml. Genotype D was predominant (98%), with viral groups D/ayw2 (73%) and D/ayw3 (27%). Group A/adw accounted for 1% of cases. Genotypes B and C were found exclusively in the Chinese immigrants (1%). Single or multiple point mutations were found in 35 cases (26%). Some of the most common mutations occurred at amino acid positions 129, 133, 134, 144, 145, including the “vaccine escape” mutation G145R. Mutations analysis revealed that amino acid substitutions did not affect detection by commercial immunoassays. J. Med. Virol. 83:245–252, 2011. © 2010 Wiley-Liss, Inc.
INTRODUCTION
Chronic hepatitis B virus (HBV) infection is an important public health problem, despite the availability of a vaccine. Worldwide, according to the World Health Organization, 360 million people suffer from chronic HBV infection [Shepard et al., 2006], with an estimation of 600,000 deaths yearly due to the acute or chronic consequences of HBV infection [Lok, 2002]. In Greece, HBV infection is considered a low to moderate endemic disease and HBV genotype surface antigen (HBsAg) carrier prevalence ranges from 0.29% to 2.6% [Raptopoulou et al., 2009].
HBV is a partially double-stranded DNA virus of the family Hepadnaviridae with a genome of 3.2 kilobases (kb) containing four partially overlapping open-reading frames encoding the polymerase (P), core (C), HBsAg, and X proteins. Based on the antigenic determinants of the HBsAg, HBV was classified into nine serological types, adw2, adw4, adr, adrq-, ayw1, ayw2, ayw3, ayw4, and ayr [Couroucé-Pauty et al., 1978]. HBsAg contains the “a” determinant (amino acid residues 124 and 147), which is common to all the HBV genotypes [Weber, 2004; Lada et al., 2006].
The heterogenicity of HBV serotype (based on single amino acid substitutions of the HBsAg), might have a positive selective value which can act as an immune evasion strategy [Simmonds, 2001]. Based on complete genome nucleotide sequence divergence of more than 8%, eight naturally occurring HBV genotypes (A–H) have been described [Norder et al., 2004; Kurbanov et al., 2010]. There is a correlation between genotypes and antigenic subtypes, with a genotype associated with two or more different subtypes. Furthermore, subgenotypes have been identified within specific genotypes, based on more than 4% sequence divergence over the entire genome [Norder et al., 2004; Kurbanov et al., 2010].
The prevalence of genotypes varies geographically, with genotype D being more common in the Mediterranean region. Subgenotypes within each genotype show distinct geographic distribution, except for those within genotype D which are found throughout the world [Norder et al., 2004].
Information concerning the genotype distribution of HBV in Greece is very limited [Katsoulidou et al., 2009]. In order to provide data on current HBV molecular epidemiology, the distribution of HBV genotypes, HBsAg subtypes and the mutations in the “a” determinant region, a retrospective study was conducted (years 2000–2007) among patients with chronic HBV infection in Thessaloniki, the second largest city in Greece. HBV genotypes were studied by partial sequencing of the surface gene, containing the HBsAg “a” antigenic determinant.
MATERIALS AND METHODS
Study Population – Source of HBV-DNA
This was an 8-year retrospective analysis of samples of 135 chronic HBV carriers admitted to Papageorgiou Regional General Hospital of Thessaloniki during the period from December 2000 until January 2007 for routine HBV-DNA detection and quantitation. All patients, all their parents/guardians in the cases of children signed an informed consent for the protocol study, which was approved by the Local Ethics committee. Socio-demographic characteristics (gender, place of birth, place of residence, and age) and medical history were recorded. Patients with HCV or HIV co-infection were excluded from the study.
Serological Assays
Routine alanine aminotransferase (ALT) and aspartate aminotransferase (AST) determinations were performed (normal range 10–37 U/L for both enzymes). The HBsAg, anti-HBc (total), anti-HBc IgM, HBeAg, and anti-HBe were determined by a commercial enzyme immunoassay method (Abbott Laboratories, Dallas, TX).
HBV-DNA Quantitative Analysis
The PCR-based assay COBAS-AMPLICOR (Roche Molecular Diagnostics, Indianapolis, IN), was used for serum HBV-DNA quantitation according to the protocol of the manufacturer [DiDomenico et al., 1996; Noborg et al., 1999]. The HBV-DNA quantitation ranges from 60 to 38,000 IU/ml (300–200,000 HBV-DNA copies/ml).
PCR Amplification of HBV-DNA S Gene
DNA was extracted from 200 µl of each serum sample by using QIAamp DNA Blood Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. The HBV-DNA sequence of 259 nucleotide pairs (329–587) from the HBsAg region, was amplified by PCR with primers (a) HB1 (5′-CAA GGT ATG TTG CCC GTT TGT-3′) and (b) HB2 (5′-AAA GCC CTG CGA ACC ACT GAA-3′). The amplified region corresponds to the amino acid residues 101–186 of the S antigen chain which contains the a-determinant region (amino acid 124–147). PCR products were separated by agarose gel electrophoresis. The clean-up of PCR products was performed according to the QIAquick PCR Purification Kit Protocol (Qiagen GmbH) using a microcentrifuge and at least 20 ng/µl of viral DNA was present in the purified product.
Direct Sequencing of PCR Products
The purified HBV-DNA was sequenced directly by Qiagen Sequencing Services using an ABI 3700 DNA automated sequencer (Applied Biosystems, Foster City, CA). All sequences were analyzed in both, forward and reverse directions. The nucleotide sequence data reported in this study has been submitted to the DDBJ/EMBL/GenBank under accession numbers: FJ178438–FJ178573.
Determination of HBsAg Subtypes
The nucleotide sequences were translated into amino acid sequences according to the open-reading frames of the partial S gene and the HBsAg subtypes were predicted from the amino acids at positions 122 (K for d and R for y determinants), 127 (P for w1–2, T for w3 and L-I for w4) and 160 (K for w and R for r) [Magnius and Norder, 1995]. Discrimination between ayw1 and awy2 was based on positions 134 and 159 (F and A, for ayw1 and Y and G for ayw2, respectively) [Norder et al., 1992].
HBV Genotypes Determination and Phylogenetic Analysis
HBV genotype was determined by sequence analysis of the 195 bp fragment from the HBV “a” determinant, using the genotyping tool available at the National Library of Medicine's National Center for Biotechnology Information (NCBI) [Rozanov et al., 2004]. In addition, HBV genotype of each isolate was deduced by comparison of the partial surface protein sequences with the sequences of the corresponding genomic regions of all HBV strains available in the GenBANK/EMBL/DDBJ database by using the BLAST program, version 2.2.8 [Altschul et al., 1990, 1997]. A phylogenetic analysis was carried out by pairwise comparison of the partial S gene sequences of representative HBV strains for all genotypes from the GenBANK/EMBL/DDBJ database with the 135 sequences obtained from the sera of the patients with chronic HBV infection, by using ClustalW [Thompson et al., 1994]. Construction of the phylogenetic tree was carried out with ClustalX (version 1.83) by using the alignment file, obtained by analysis with ClustalW. By using this program the distances between all pairs of sequences were calculated, and this was followed by application of the neighbor-joining method to the distance matrix. Confidence values for the groups in the tree (bootstrap values on a scale from 1 to 1,000) were also calculated by using ClustalX. Dendrograms showing the phylogenetic relationships among the HBV isolates of the present study and prototype strains was plotted in the PHYLIP format output by using the TreeView software (version 3.0), which was obtained from the website of the University of Glasgow.
Statistics
Statistical analysis of data was performed using the Statistical Program for Social Sciences (SPSS version 16.0). Results were presented as mean values ± standard deviation (SD). Comparisons between patients groups were performed using the students t-test or nonparametric Mann–Whitney U-test. A P value <0.05 was considered significant.
RESULTS
HBV Serology and DNA Viral Load
A total of 135 HBV carriers (93 males and 42 females) with ages ranging from 9 to 92 years (mean value 45.5 years), were enrolled in the study (Table I). Of the 135 patients, 124 patients were ethnic Greek residents of Thessaloniki (Northern Greece) while the rest were immigrants from Albania (five patients), China (two patients), Turkey (two patients), and Georgia (two patients).
| Number of patients | Genotypes/subtypes (no. patients) | |
|---|---|---|
| Gender (male/female) | 135 (93/42) | |
| Age (mean years) (range) | 45.5 (9–92) | |
| Origin | ||
| Greece (GR) | 124 | D/ayw3 (30) |
| D/ayw2 (93) | ||
| A2/ayw2 (1) | ||
| Albania(Al) | 5 | D/ayw3 (4) |
| D/ayw2 (1) | ||
| China (Ch) | 2 | C/adr(1), B/Nd |
| Georgia (GeO) | 2 | D/ayw3 (2) |
| Turkey (TU) | 2 | D/ayw3 (1) |
| D/ayw2 (1) | ||
- Patients sequences are reported in DDBJ/EMBL/GenBank under accession numbers FJ178438–FJ178573). Nd, not determined.
All patients with chronic HBV infection had elevated aminotransferase levels. Mean ALT and AST levels were 238 IU/L (SD = 576.84) and 152 IU/L (SD = 282.65), respectively. All patients were HBsAg positive, and the majority of them (62.5%; 84/135) were anti-HBe positive. Thirty-five per cent (47/135) of patients were HbeAg positive, while 3% (4/135) were both HbeAg positive and anti-HBe positive. Anti-HBc seropositivity was detected in 98% of the study population. In 4.4% (6/135) of patients, simultaneous presence of HBsAg and anti-HBs was detected. Serum HBV-DNA levels ranged from 1.02 × 105 to 2.2 × 107 IU/ml. Patients positive for both HBeAg and anti-HBe, had significantly higher DNA levels than HBeAg negative patients (2.29 × 106 vs. 1.3 × 106; P < 0.01).
Determination of HBV Genotypes
The results obtained by analysis of the nucleotide sequences from the 135 serum samples tested, are summarized in TablesII–III and the phylogenetic trees in Figures 1 and 2. The sequences grouped within clusters corresponding to genotypes A, B, C, and D of HBV. None of them grouped within genotypes E, F, G, or H. All genotype clusters were supported by significant bootstrap values. Agreement between the plylogenetic analysis and the NCBI genotyping tool results was observed in all cases and an HBV genotype was assigned to all but one samples. The distribution of HBV genotypes found among the 135 serum samples was 98% for genotype D, which was almost exclusively prevalent, 1% for genotype A, and 0.5% for each genotype B and C, which were exclusively found in Asian immigrants. Recombination of the HBV genes from different genotypes was not observed. The sequences of the partial S gene from the 135 patients participated in the study are available at the GenBank/EMBL/DDBJ database with accession numbers: FJ17843–FJ178573.
| HBsAg subtype | Genotype | |||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | Total | |
| ayw2 | — | — | — | 96 | — | — | — | 96 |
| ayw3 | — | — | — | 36 | — | — | — | 36 |
| adw2 | 1 | — | — | — | — | — | — | 1 |
| Adr | — | — | 1 | — | — | — | — | 1 |
| No subtype | — | 1 | — | — | — | — | — | 1 |
| Total | 1 | 1 | 1 | 132 | — | — | — | 135 |
| HBV serology profile | Total no (%) | ayw2 | ayw3 | Mutations | HBV-DNA (IU/ml) | ALT (IU/ml) |
|---|---|---|---|---|---|---|
| HBsAg+/HBeAb+ | 82 (61%) | 58 (71%) | 24 (29%) | ayw2 (15%), ayw3 (42%) | ||
| HBV829GR | A128L | 3,260,000 | 2,610 | |||
| HBV1010GR | A128V | 7,070,000 | 88 | |||
| HBV548GR | A128V | 206,000 | 674 | |||
| HBV641GR | D144E | 2,430,000 | 30 | |||
| HBV998GR | E164G, S174N, S167L | 3,760,000 | 57 | |||
| HBV205GR | F158L | 925,000 | 58 | |||
| HBV275GR | M133T | 2,260,000 | 41 | |||
| HBV400GR | M133T | 580,000 | 42 | |||
| HBV895GR | M133T | 1,870,000 | 77 | |||
| HBV1059GR | P127A, Q129R | 1,750,000 | 240 | |||
| HBV1387GR | P142T, D144V | 943,000 | 1,147 | |||
| HBV868GR | Q129G | 12,200,000 | 393 | |||
| HBV552GR | Q129H,G130R | 7,660,000 | 108 | |||
| HBV1141GR | S174N | 1,870,000 | 1,095 | |||
| HBV204GR | T126S | 436,000 | 215 | |||
| HBV982GR | T131I | 3,420,000 | 85 | |||
| HBV785GR | Y134H | 8,400,000 | 132 | |||
| HBV863GR | Y134N | 818,000 | 560 | |||
| HBV1586GR | Y134N | 912,000 | 289 | |||
| HBsAg+/HBeAg+* | 43* (31.6%) | 29* (67%) | 11* (26%) | ayw2 (22%), ayw3 (27%) | ||
| HBV1010GR | A128V | 4,550,000 | ||||
| HBV928GR | E164V | 13,000,000 | ||||
| HBV1002GR | E164V | 7,330,000 | ||||
| HBV1090GR | G145A | 1,370,000 | ||||
| HBV690GR** | I126T | NT | ||||
| HBV1696GR | P127L | 1,200,000 | ||||
| HBV262GR | Q129H | 2,740,000 | ||||
| HBV1642GR | Q129R | 7,700,000 | ||||
| HBV742GR | S136F | NT | ||||
| HBV1774GR | V177L | 8,900,000 | ||||
| HBV1586GR | Y134N | 912,000 | ||||
| HBsAg+/HBeAg+/HBeAb+ | 3 (2.2%) | 3 | — | |||
| HBV595GR | S174H | 11,000,000 | 107 | |||
| HBsAg+/HBcAb-/HBeAg+ | 1 (0.7%) | 1 | — | — | ||
| HBsAg+/HBsAb+ | 5(3.4%) | 4 (80%) | 1 (20%) | |||
| HBV804GR | T123N,M133L,G145R,V177A | NT | 39 | |||
| HBV420GR | D144E,S143L,P142L,S136F,G145R | 1,260,000 | 6 | |||
| HBV347GR | S136F | 2,120,000 | 24 | |||
| HBsAg+/HBsAb+/HBeAg+/HBeAb+ | 1 (0.7%) | 1 | ||||
| Total | 135* | 96* | 36* |
- *In the HBsAg+/HBeAg+ group there are three patients with different subtypes than ayw2 or ayw3, one with adr (**) subtype with mutation I126T, the second has adw2 subtype and the third one has no detectable subtype.
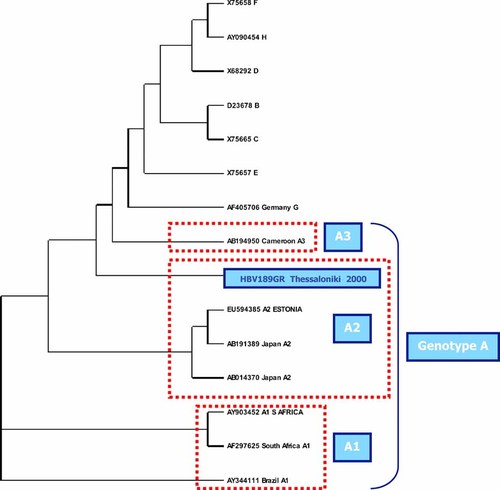
Phylogenetic tree of the HBV isolate from genotype A.
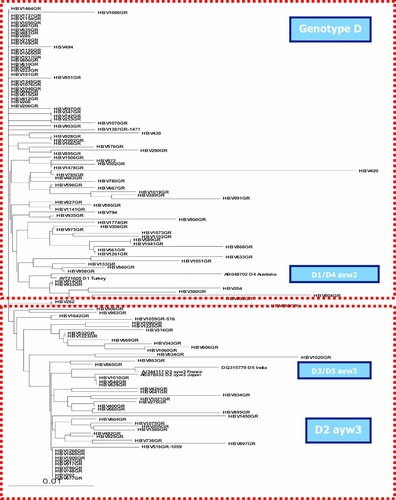
Phylogenetic tree of all HBV isolates from genotype D.
Determination of HBsAg Subtypes and Association With HBV Genotypes
Partial sequencing of the S gene predicted the HBsAg subtypes in all but one case. Isolates from genotype D specified exclusively subtype ayw2 (96/132, 73%) and subtype ayw3 (36/132, 27%). Almost all these isolates grouped together in HBsAg subtypes in the phylogenetic tree (Fig. 2). The only strain belonging to genotype A specified subtype adw2, was isolated from a chronic HBV native Greek carrier. In addition, the one strain belonging to genotype C was detected in a Chinese immigrant and belonged to subtype adr. The second strain detected from another immigrant from China belonged to genotype B, but the HBsAg subtype was not specified, as the HBsAg aminoacid sequence was only readable between positions 121 and 154. The first part of the sequence seemed to specify subtype ayw (Thr126), while the second part seemed to specify subtype adw (Thr143).
No unusual genotype/HBsAg subtype combinations were found. The association of HBsAg subtypes with genotypes and seroconvertion status is shown in Tables II and III, respectively. The ayw2 HBsAg subtype was predominant in both the HBeAg positive and the anti-HBe positive patients with 74% (32/43) and 71% (58/82), respectively. The ayw3 subtype was less common with 26% (11/43) in the HBeAg positive patients and 29% (24/82) in the anti-HBe positive patients.
HBV Genetic Diversity
Single or multiple point mutations were found in 26% (35/135) of the cases as shown in Tables I and III. Some of the most common mutations occurred at amino acid positions 129 and 134 (five strains), 133 and 145 (four strains, including the “vaccine escape” mutant G145R), and 128, 144, 164, 174 (three strains each). Most common mutations observed were M133T, Y134N, and S174N (3 isolates). In the anti-HBe positive group, the mutation rate was significantly higher in patients with the ayw3 subtype compared to the ayw2 group (42% vs. 15%; P < 0.01). Such difference was not observed in the HBeAg positive group (27% vs. 22%; P > 0.01). Multiple point mutations were almost exclusively observed in the anti-HBe positive group and in two of the four patients belonging to the HBsAg positive/anti-HBs positive group (Fig. 3). Most strains were specified as subtype ayw2. Interestingly, 5- and 4-point mutations were found in the two patients from the HBsAg positive/anti-HBs positive group (Fig. 3). In the first case, an HBeAg positive leukemic patient under chemotherapy and immunosuppression, the point mutations were S136F, P142L, S143L, D144E, and the “vaccine escape” mutation G145R. In the latter case, an anti-HBe positive chronic HBV carrier, the point mutations were T123N, M133L, G145R, and V177A.

Comparison with reference sequences for mutant isolates (a) HBV804GR and (b) HBV420GR. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com]
DISCUSSION
In this 8-year retrospective study, important data were obtained on the HBV genotypes/subtypes and mutations in HBsAg in chronic HBV carriers in Thessaloniki, Greece. Two HBV genotypes A and D, were found among native Greeks, with genotype D being almost prevalent exclusively (D: 98% vs. A: 1%). Genotype D was also the only genotype found in Turkey [Bozdayi et al., 2005; Ozaslan et al., 2007] and the predominant genotype in Serbia [Lazarevic et al., 2007].
The remaining 1% derived from two HBV strains from Chinese immigrants, were specified as genotypes B and C. The one isolate belonging to genotype A in this study was grouped with a reference sequence for A2, suggesting a European origin. As expected for samples from the European population, this strain belongs to subtype adw2.
The presence of both HBsAg and anti-HBs, was found in 4% of the study population. The selection of HBsAg variants is a possible mechanism underlying the presence of both HBsAg and anti-HBs. Residue changes in the “a” determinant of the S protein, the main target of anti-HBs antibodies, could lead to escape from recognition by the host immune system. However, HBV escape mutants may also occur naturally in chronic HBV carriers due to the exclusive pressure of the host immune system. The results of the present study are comparable to those reported by Yamamoto et al. [1994], Kohno et al. [1996], and Lada et al. [2006].
The direct sequencing of a relatively short fragment of the HBV genome was shown to be efficient for the determination of both viral genotype and HBsAg subtype, as well as mutations with potential impact on the HBsAg antigenicity. Mutational studies on HBV S genes are important in understanding the failure of protection with current HBV vaccines and the conflicting results of HBsAg detection with different diagnostic kits. This can be due to the fact that the specificity of the antibodies used in diagnostic assays and vaccines are targeted to these regions, and especially the “a” determinant. The results from this study, confirmed the good performance of the tests used, even in the presence of multiple point mutations.
In conclusion, data from this study contribute to the investigation of HBV genotype and HBsAg subtype distribution in Greece. Partial S gene sequence analysis determined that the HBV genotype D and ayw2 HBsAg subtype, were predominant in patients with chronic HBV infection from northern Greece. The clustering of all DNA sequences into genotype groups corresponded to their respective HBsAg subtype. Analysis of the point mutations revealed that amino acid substitutions at immunodominant epitopes involved in B- or/and T-cell recognition did not affect detection by commercial immunoassays. Additional data will be required in order to determine the frequency of HBV genotypes in Greece.
Acknowledgements
The authors would like to thank all medical doctors and technicians working in the Virology Laboratory, Papageorgiou Regional General Hospital, Thessaloniki, Greece, who assisted in the study. The authors would also like to acknowledge Abbott Hellas for financing the S gene partial sequencing of the HBV strains.




